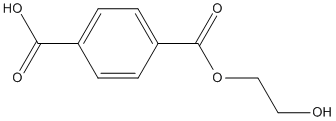
MHET
product of hydrolysis of Polyethylene-terephthalate or HEMT model substrate. Polyethylene terephthalate is the most common thermoplastic polymer resin of the polyester family and is used in fibers for clothing, containers for liquids and foods. Degradation of PET using hydrolytic enzymes leads to the release of hydrolysis products such as BHET (bis(2-hydroxyethyl) terephthalate), MHET (mono-(2-hydroxyethyl) terephthalic acid), TPA (terephthalic acid) and EG (ethylene glycol)
General
Type : Terephthalate,Phthalate,Benzoate
Chemical_Nomenclature : 4-(2-hydroxyethoxycarbonyl)benzoic acid
Canonical SMILES : C1=CC(=CC=C1C(=O)O)C(=O)OCCO
InChI : InChI=1S\/C10H10O5\/c11-5-6-15-10(14)8-3-1-7(2-4-8)9(12)13\/h1-4,11H,5-6H2,(H,12,13)
InChIKey : BCBHDSLDGBIFIX-UHFFFAOYSA-N
Other name(s) : 4-((2-Hydroxyethoxy)carbonyl)benzoic acid,Mono(ethylene terephthalate),Ethylene glycol, cyclic ester with (C7-C11)dibasic acids,AC1L5ATF,ET,C9C,4-[(2-hydroxyethoxy)carbonyl]benzoate,1,4-Benzenedicarboxylic acid, mono(2-hydroxyethyl) ester,CTK0G1127,CHEBI:131701,CHEBI:131704
MW : 210.18
Formula : C10H10O5
CAS_number : 1137-99-1
UniChem : BCBHDSLDGBIFIX-UHFFFAOYSA-N
IUPHAR :
Wikipedia :
Target
Families : MHET ligand of proteins in family: Polyesterase-lipase-cutinase || Tannase
Stucture : 7W1I Crystal structure of carboxylesterase mutant from Thermobifida fusca with C8X (BHET) and C9C (MHET)
Protein : idesa-peth || idesa-mheth || thefu-q6a0i3 || thefu-q6a0i4 || 9bact-g9by57
References (11)
| Title : Complete bio-degradation of poly(butylene adipate-co-terephthalate) via engineered cutinases - Yang_2023_Nat.Commun_14_1645 |
| Author(s) : Yang Y , Min J , Xue T , Jiang P , Liu X , Peng R , Huang JW , Qu Y , Li X , Ma N , Tsai FC , Dai L , Zhang Q , Liu Y , Chen CC , Guo RT |
| Ref : Nat Commun , 14 :1645 , 2023 |
| Abstract : Yang_2023_Nat.Commun_14_1645 |
| ESTHER : Yang_2023_Nat.Commun_14_1645 |
| PubMedSearch : Yang_2023_Nat.Commun_14_1645 |
| PubMedID: 36964144 |
| Gene_locus related to this paper: idesa-peth , thefu-q6a0i4 |
| Title : Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy - Zeng_2022_ACS.Catal_12_3033 |
| Author(s) : Zeng W , Li X , Yang Y , Min J , Huang JW , Liu W , Niu D , Yang X , Han X , Zhang L , Dai L , Chen CC , Guo RT |
| Ref : ACS Catal , 12 :3033 , 2022 |
| Abstract : Zeng_2022_ACS.Catal_12_3033 |
| ESTHER : Zeng_2022_ACS.Catal_12_3033 |
| PubMedSearch : Zeng_2022_ACS.Catal_12_3033 |
| PubMedID: |
| Gene_locus related to this paper: 9bact-g9by57 |
| Title : Novel efficient enzymatic synthesis of the key-reaction intermediate of PET depolymerization, mono(2-hydroxyethyl terephthalate) - MHET - de Queiros Eugenio_2022_J.Biotechnol__ |
| Author(s) : de Queiros Eugenio E , Campisano ISP , Dias AG , de Castro AM , Coelho MAZ , Langone MAP |
| Ref : J Biotechnol , : , 2022 |
| Abstract : de Queiros Eugenio_2022_J.Biotechnol__ |
| ESTHER : de Queiros Eugenio_2022_J.Biotechnol__ |
| PubMedSearch : de Queiros Eugenio_2022_J.Biotechnol__ |
| PubMedID: 36063976 |
| Gene_locus related to this paper: humin-cut |
| Title : Structure-function analysis of two closely related cutinases from Thermobifida cellulosilytica - Baath_2021_Biotechnol.Bioeng__ |
| Author(s) : Baath JA , Novy V , Carneiro LV , Guebitz GM , Olsson L , Westh P , Ribitsch D |
| Ref : Biotechnol Bioeng , : , 2021 |
| Abstract : Baath_2021_Biotechnol.Bioeng__ |
| ESTHER : Baath_2021_Biotechnol.Bioeng__ |
| PubMedSearch : Baath_2021_Biotechnol.Bioeng__ |
| PubMedID: 34755331 |
| Gene_locus related to this paper: thefu-q6a0i4 , thefu-q6a0i3 |
| Title : Active Site Flexibility as a Hallmark for Efficient PET Degradation by I. sakaiensis PETase - Fecker_2018_Biophys.J_114_1302 |
| Author(s) : Fecker T , Galaz-Davison P , Engelberger F , Narui Y , Sotomayor M , Parra LP , Ramirez-Sarmiento CA |
| Ref : Biophysical Journal , 114 :1302 , 2018 |
| Abstract : Fecker_2018_Biophys.J_114_1302 |
| ESTHER : Fecker_2018_Biophys.J_114_1302 |
| PubMedSearch : Fecker_2018_Biophys.J_114_1302 |
| PubMedID: 29590588 |
| Gene_locus related to this paper: idesa-peth |
| Title : Characterization and engineering of a plastic-degrading aromatic polyesterase - Austin_2018_Proc.Natl.Acad.Sci.U.S.A_115_E4350 |
| Author(s) : Austin HP , Allen MD , Donohoe BS , Rorrer NA , Kearns FL , Silveira RL , Pollard BC , Dominick G , Duman R , El Omari K , Mykhaylyk V , Wagner A , Michener WE , Amore A , Skaf MS , Crowley MF , Thorne AW , Johnson CW , Woodcock HL , McGeehan JE , Beckham GT |
| Ref : Proc Natl Acad Sci U S A , 115 :E4350 , 2018 |
| Abstract : Austin_2018_Proc.Natl.Acad.Sci.U.S.A_115_E4350 |
| ESTHER : Austin_2018_Proc.Natl.Acad.Sci.U.S.A_115_E4350 |
| PubMedSearch : Austin_2018_Proc.Natl.Acad.Sci.U.S.A_115_E4350 |
| PubMedID: 29666242 |
| Gene_locus related to this paper: idesa-peth |
| Title : Structural insight into molecular mechanism of poly(ethylene terephthalate) degradation - Joo_2018_Nat.Commun_9_382 |
| Author(s) : Joo S , Cho IJ , Seo H , Son HF , Sagong HY , Shin TJ , Choi SY , Lee SY , Kim KJ |
| Ref : Nat Commun , 9 :382 , 2018 |
| Abstract : Joo_2018_Nat.Commun_9_382 |
| ESTHER : Joo_2018_Nat.Commun_9_382 |
| PubMedSearch : Joo_2018_Nat.Commun_9_382 |
| PubMedID: 29374183 |
| Gene_locus related to this paper: idesa-peth |
| Title : New insights into the function and global distribution of polyethylene terephthalate (PET) degrading bacteria and enzymes in marine and terrestrial metagenomes - Danso_2018_Appl.Environ.Microbiol_84_e2773 |
| Author(s) : Danso D , Schmeisser C , Chow J , Zimmermann W , Wei R , Leggewie C , Li X , Hazen T , Streit WR |
| Ref : Applied Environmental Microbiology , 84 :e2773 , 2018 |
| Abstract : Danso_2018_Appl.Environ.Microbiol_84_e2773 |
| ESTHER : Danso_2018_Appl.Environ.Microbiol_84_e2773 |
| PubMedSearch : Danso_2018_Appl.Environ.Microbiol_84_e2773 |
| PubMedID: 29427431 |
| Gene_locus related to this paper: 9burk-PET10 , 9burk-PET11 , 9gamm-a0a0d4l7e6 , 9alte-n6vy44 , 9zzzz-a0a0f9x315 , deiml-e8u721 , olean-r4ykl9 , vibga-a0a1z2siq1 , 9burk-a0a0g3bi90 , 9bact-c3ryl0 , 9actn-h6wx58 , idesa-peth , 9bact-g9by57 , acide-PBSA , morsp-lip1 |
| Title : Structural insight into catalytic mechanism of PET hydrolase - Han_2017_Nat.Commun_8_2106 |
| Author(s) : Han X , Liu W , Huang JW , Ma J , Zheng Y , Ko TP , Xu L , Cheng YS , Chen CC , Guo RT |
| Ref : Nat Commun , 8 :2106 , 2017 |
| Abstract : Han_2017_Nat.Commun_8_2106 |
| ESTHER : Han_2017_Nat.Commun_8_2106 |
| PubMedSearch : Han_2017_Nat.Commun_8_2106 |
| PubMedID: 29235460 |
| Gene_locus related to this paper: idesa-peth |
| Title : A bacterium that degrades and assimilates poly(ethylene terephthalate) - Yoshida_2016_Science_351_1196 |
| Author(s) : Yoshida S , Hiraga K , Takehana T , Taniguchi I , Yamaji H , Maeda Y , Toyohara K , Miyamoto K , Kimura Y , Oda K |
| Ref : Science , 351 :1196 , 2016 |
| Abstract : Yoshida_2016_Science_351_1196 |
| ESTHER : Yoshida_2016_Science_351_1196 |
| PubMedSearch : Yoshida_2016_Science_351_1196 |
| PubMedID: 26965627 |
| Gene_locus related to this paper: idesa-mheth , idesa-peth |
| Title : Crystal Structure and Thermodynamic and Kinetic Stability of Metagenome-Derived LC-Cutinase - Sulaiman_2014_Biochemistry_53_1858 |
| Author(s) : Sulaiman S , You DJ , Kanaya E , Koga Y , Kanaya S |
| Ref : Biochemistry , 53 :1858 , 2014 |
| Abstract : Sulaiman_2014_Biochemistry_53_1858 |
| ESTHER : Sulaiman_2014_Biochemistry_53_1858 |
| PubMedSearch : Sulaiman_2014_Biochemistry_53_1858 |
| PubMedID: 24593046 |
| Gene_locus related to this paper: 9bact-g9by57 |