BHET
General
Type : Terephthalate || Phthalate
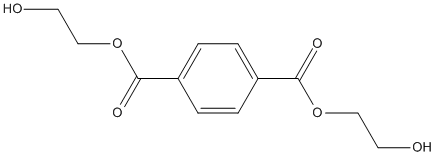
Chemical_Nomenclature : bis(2-hydroxyethyl) benzene-1,4-dicarboxylate
Canonical SMILES : C1=CC(=CC=C1C(=O)OCCO)C(=O)OCCO
InChI : InChI=1S\/C12H14O6\/c13-5-7-17-11(15)9-1-2-10(4-3-9)12(16)18-8-6-14\/h1-4,13-14H,5-8H2
InChIKey : QPKOBORKPHRBPS-UHFFFAOYSA-N
Other name(s) : bis-2(hydroxyethyl) terephthalate, Bis(hydroxyethyl) terephthalate, Bis(beta-hydroxyethyl) terephthalate, 1,4-Benzenedicarboxylic acid, bis(2-hydroxyethyl) ester, Terephthalic acid diethylene glycol ester, Bis(ethylene glycol) terephthalate, Bis(2-hydroxyethyl) terephthalate, bis(2 hydroxyethyl) terephthalate, ETE, C8X, 2HE(MHET), HETPAL01
References (20)
| Title : An archaeal lid-containing feruloyl esterase degrades polyethylene terephthalate - Perez-Garcia_2023_Commun.Chem_6_193 |
| Author(s) : Perez-Garcia P , Chow J , Costanzi E , Gurschke M , Dittrich J , Dierkes RF , Molitor R , Applegate V , Feuerriegel G , Tete P , Danso D , Thies S , Schumacher J , Pfleger C , Jaeger KE , Gohlke H , Smits SHJ , Schmitz RA , Streit WR |
| Ref : Commun Chem , 6 :193 , 2023 |
| Abstract : Perez-Garcia_2023_Commun.Chem_6_193 |
| ESTHER : Perez-Garcia_2023_Commun.Chem_6_193 |
| PubMedSearch : Perez-Garcia_2023_Commun.Chem_6_193 |
| PubMedID: 37697032 |
| Gene_locus related to this paper: 9arch-PETcan211 , 9cren-PETcan204 , 9arch-PET46 |
| Title : Structural and Biochemical Insights into Bis(2-hydroxyethyl) Terephthalate Degrading Carboxylesterase Isolated from Psychrotrophic Bacterium Exiguobacterium antarcticum - Hwang_2023_Int.J.Mol.Sci_24_12022 |
| Author(s) : Hwang J , Yoo W , Shin SC , Kim KK , Kim HW , Do H , Lee JH |
| Ref : Int J Mol Sci , 24 :12022 , 2023 |
| Abstract : Hwang_2023_Int.J.Mol.Sci_24_12022 |
| ESTHER : Hwang_2023_Int.J.Mol.Sci_24_12022 |
| PubMedSearch : Hwang_2023_Int.J.Mol.Sci_24_12022 |
| PubMedID: 37569396 |
| Gene_locus related to this paper: human-DPP9 , exiab-k0afs0 |
| Title : The Mobility of the Cap Domain Is Essential for the Substrate Promiscuity of a Family IV Esterase from Sorghum Rhizosphere Microbiome - Distaso_2023_Appl.Environ.Microbiol__e0180722 |
| Author(s) : Distaso M , Cea-Rama I , Coscolin C , Chernikova TN , Tran H , Ferrer M , Sanz-Aparicio J , Golyshin PN |
| Ref : Applied Environmental Microbiology , :e0180722 , 2023 |
| Abstract : Distaso_2023_Appl.Environ.Microbiol__e0180722 |
| ESTHER : Distaso_2023_Appl.Environ.Microbiol__e0180722 |
| PubMedSearch : Distaso_2023_Appl.Environ.Microbiol__e0180722 |
| PubMedID: 36602332 |
| Gene_locus related to this paper: 9bact-EH0 |
| Title : Discovery and mechanism-guided engineering of BHET hydrolases for improved PET recycling and upcycling - Li_2023_Nat.Commun_14_4169 |
| Author(s) : Li A , Sheng Y , Cui H , Wang M , Wu L , Song Y , Yang R , Li X , Huang H |
| Ref : Nat Commun , 14 :4169 , 2023 |
| Abstract : Li_2023_Nat.Commun_14_4169 |
| ESTHER : Li_2023_Nat.Commun_14_4169 |
| PubMedSearch : Li_2023_Nat.Commun_14_4169 |
| PubMedID: 37443360 |
| Gene_locus related to this paper: 9flao-ChryBHETase , bacsu-pnbae |
| Title : A novel Bacillus subtilis BPM12 with high bis(2 hydroxyethyl)terephthalate hydrolytic activity efficiently interacts with virgin and mechanically recycled polyethylene terephthalate - Pantelic_2023_Environ.Technol.Innov_32_103316 |
| Author(s) : Pantelic B , Araujo JA , Jeremic S , Azeem M , Attallah OA , Siaperas R , Mojicevic M , Chen Y , Brennan-Fournet M , Topakas E , Nikodinovic-Runic J |
| Ref : Environmental Technology & Innovation , 32 :103316 , 2023 |
| Abstract : Pantelic_2023_Environ.Technol.Innov_32_103316 |
| ESTHER : Pantelic_2023_Environ.Technol.Innov_32_103316 |
| PubMedSearch : Pantelic_2023_Environ.Technol.Innov_32_103316 |
| PubMedID: |
| Gene_locus related to this paper: bacsu-pnbae |
| Title : Biochemical Characterization and NMR Study of a PET-Hydrolyzing Cutinase from Fusarium solani pisi - Hellesnes_2023_Biochemistry__ |
| Author(s) : Hellesnes KN , Vijayaraj S , Fojan P , Petersen E , Courtade G |
| Ref : Biochemistry , : , 2023 |
| Abstract : Hellesnes_2023_Biochemistry__ |
| ESTHER : Hellesnes_2023_Biochemistry__ |
| PubMedSearch : Hellesnes_2023_Biochemistry__ |
| PubMedID: 36967526 |
| Gene_locus related to this paper: fusso-cutas |
| Title : Characterization of a PBAT Degradation Carboxylesterase from Thermobacillus composti KWC4 - Wu_2023_Catalysts_13_340 |
| Author(s) : Wu P , Li Z , Gao J , Zhao Y , Wang H , Qin H , Gu Q , Wei R , Liu W , Han X |
| Ref : Catalysts , 13 :340 , 2023 |
| Abstract : Wu_2023_Catalysts_13_340 |
| ESTHER : Wu_2023_Catalysts_13_340 |
| PubMedSearch : Wu_2023_Catalysts_13_340 |
| PubMedID: |
| Gene_locus related to this paper: theck-l0ef70 |
| Title : Mechanistic studies of a lipase unveil effect of pH on hydrolysis products of small PET modules - Swiderek_2023_Nat.Commun_14_3556 |
| Author(s) : Swiderek K , Velasco-Lozano S , Galmes M , Olazabal I , Sardon H , Lopez-Gallego F , Moliner V |
| Ref : Nat Commun , 14 :3556 , 2023 |
| Abstract : Swiderek_2023_Nat.Commun_14_3556 |
| ESTHER : Swiderek_2023_Nat.Commun_14_3556 |
| PubMedSearch : Swiderek_2023_Nat.Commun_14_3556 |
| PubMedID: 37321996 |
| Gene_locus related to this paper: canar-LipB |
| Title : Structural Insights into (Tere)phthalate-Ester Hydrolysis by a Carboxylesterase and Its Role in Promoting PET Depolymerization - von Haugwitz_2022_ACS.Catal_12_15259 |
| Author(s) : von Haugwitz G , Han X , Pfaff L , Li Q , Wei H , Gao J , Methling K , Ao Y , Brack Y , Jan Mican J , Feiler CG , Weiss MS , Bednar D , Palm GJ , Lalk M , Lammers M , Damborsky J , Weber G , Liu W , Bornscheuer UT , Wei R |
| Ref : ACS Catal , 12 :15259 , 2022 |
| Abstract : von Haugwitz_2022_ACS.Catal_12_15259 |
| ESTHER : von Haugwitz_2022_ACS.Catal_12_15259 |
| PubMedSearch : von Haugwitz_2022_ACS.Catal_12_15259 |
| PubMedID: 36570084 |
| Gene_locus related to this paper: thefu-1831 |
| Title : Structure-function analysis of two closely related cutinases from Thermobifida cellulosilytica - Baath_2021_Biotechnol.Bioeng__ |
| Author(s) : Baath JA , Novy V , Carneiro LV , Guebitz GM , Olsson L , Westh P , Ribitsch D |
| Ref : Biotechnol Bioeng , : , 2021 |
| Abstract : Baath_2021_Biotechnol.Bioeng__ |
| ESTHER : Baath_2021_Biotechnol.Bioeng__ |
| PubMedSearch : Baath_2021_Biotechnol.Bioeng__ |
| PubMedID: 34755331 |
| Gene_locus related to this paper: thefu-q6a0i4 , thefu-q6a0i3 |
| Title : The Bacteroidetes Aequorivita sp. and Kaistella jeonii Produce Promiscuous Esterases With PET-Hydrolyzing Activity - Zhang_2022_Front.Microbiol_12_803896 |
| Author(s) : Zhang H , Perez-Garcia P , Dierkes RF , Applegate V , Schumacher J , Chibani CM , Sternagel S , Preuss L , Weigert S , Schmeisser C , Danso D , Pleiss J , Almeida A , Hocker B , Hallam SJ , Schmitz RA , Smits SHJ , Chow J , Streit WR |
| Ref : Front Microbiol , 12 :803896 , 2021 |
| Abstract : Zhang_2022_Front.Microbiol_12_803896 |
| ESTHER : Zhang_2022_Front.Microbiol_12_803896 |
| PubMedSearch : Zhang_2022_Front.Microbiol_12_803896 |
| PubMedID: 35069509 |
| Gene_locus related to this paper: flutr-f2ie04 , 9flao-a0a0c1f4u8 , 9flao-kjj39608 , 9flao-a0a330mq60 |
| Title : Decomposition of the PET Film by MHETase Using Exo-PETase Function - Sagong_2020_ACS.Catal_10_48050 |
| Author(s) : Sagong HY , Seo H , Kim T , Son H , Joo S , Lee S , Kim S , Woo JS , Hwang S , Kim KJ |
| Ref : ACS Catal , 10 :4805 , 2020 |
| Abstract : Sagong_2020_ACS.Catal_10_48050 |
| ESTHER : Sagong_2020_ACS.Catal_10_48050 |
| PubMedSearch : Sagong_2020_ACS.Catal_10_48050 |
| PubMedID: |
| Gene_locus related to this paper: idesa-mheth |
| Title : Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate - Palm_2019_Nat.Commun_10_1717 |
| Author(s) : Palm GJ , Reisky L , Bottcher D , Muller H , Michels EAP , Walczak MC , Berndt L , Weiss MS , Bornscheuer UT , Weber G |
| Ref : Nat Commun , 10 :1717 , 2019 |
| Abstract : Palm_2019_Nat.Commun_10_1717 |
| ESTHER : Palm_2019_Nat.Commun_10_1717 |
| PubMedSearch : Palm_2019_Nat.Commun_10_1717 |
| PubMedID: 30979881 |
| Gene_locus related to this paper: idesa-mheth |
| Title : Active Site Flexibility as a Hallmark for Efficient PET Degradation by I. sakaiensis PETase - Fecker_2018_Biophys.J_114_1302 |
| Author(s) : Fecker T , Galaz-Davison P , Engelberger F , Narui Y , Sotomayor M , Parra LP , Ramirez-Sarmiento CA |
| Ref : Biophysical Journal , 114 :1302 , 2018 |
| Abstract : Fecker_2018_Biophys.J_114_1302 |
| ESTHER : Fecker_2018_Biophys.J_114_1302 |
| PubMedSearch : Fecker_2018_Biophys.J_114_1302 |
| PubMedID: 29590588 |
| Gene_locus related to this paper: idesa-peth |
| Title : Characterization and engineering of a plastic-degrading aromatic polyesterase - Austin_2018_Proc.Natl.Acad.Sci.U.S.A_115_E4350 |
| Author(s) : Austin HP , Allen MD , Donohoe BS , Rorrer NA , Kearns FL , Silveira RL , Pollard BC , Dominick G , Duman R , El Omari K , Mykhaylyk V , Wagner A , Michener WE , Amore A , Skaf MS , Crowley MF , Thorne AW , Johnson CW , Woodcock HL , McGeehan JE , Beckham GT |
| Ref : Proc Natl Acad Sci U S A , 115 :E4350 , 2018 |
| Abstract : Austin_2018_Proc.Natl.Acad.Sci.U.S.A_115_E4350 |
| ESTHER : Austin_2018_Proc.Natl.Acad.Sci.U.S.A_115_E4350 |
| PubMedSearch : Austin_2018_Proc.Natl.Acad.Sci.U.S.A_115_E4350 |
| PubMedID: 29666242 |
| Gene_locus related to this paper: idesa-peth |
| Title : Structural insight into molecular mechanism of poly(ethylene terephthalate) degradation - Joo_2018_Nat.Commun_9_382 |
| Author(s) : Joo S , Cho IJ , Seo H , Son HF , Sagong HY , Shin TJ , Choi SY , Lee SY , Kim KJ |
| Ref : Nat Commun , 9 :382 , 2018 |
| Abstract : Joo_2018_Nat.Commun_9_382 |
| ESTHER : Joo_2018_Nat.Commun_9_382 |
| PubMedSearch : Joo_2018_Nat.Commun_9_382 |
| PubMedID: 29374183 |
| Gene_locus related to this paper: idesa-peth |
| Title : Fast turbidimetric assay for analyzing the enzymatic hydrolysis of polyethylene terephthalate model substrates - Belisario-Ferrari_2018_Biotechnol.J__e1800272 |
| Author(s) : Belisario-Ferrari MR , Wei R , Schneider T , Honak A , Zimmermann W |
| Ref : Biotechnol J , :e1800272 , 2018 |
| Abstract : Belisario-Ferrari_2018_Biotechnol.J__e1800272 |
| ESTHER : Belisario-Ferrari_2018_Biotechnol.J__e1800272 |
| PubMedSearch : Belisario-Ferrari_2018_Biotechnol.J__e1800272 |
| PubMedID: 30430764 |
| Gene_locus related to this paper: thefu-1831 |
| Title : New insights into the function and global distribution of polyethylene terephthalate (PET) degrading bacteria and enzymes in marine and terrestrial metagenomes - Danso_2018_Appl.Environ.Microbiol_84_e2773 |
| Author(s) : Danso D , Schmeisser C , Chow J , Zimmermann W , Wei R , Leggewie C , Li X , Hazen T , Streit WR |
| Ref : Applied Environmental Microbiology , 84 :e2773 , 2018 |
| Abstract : Danso_2018_Appl.Environ.Microbiol_84_e2773 |
| ESTHER : Danso_2018_Appl.Environ.Microbiol_84_e2773 |
| PubMedSearch : Danso_2018_Appl.Environ.Microbiol_84_e2773 |
| PubMedID: 29427431 |
| Gene_locus related to this paper: 9burk-PET10 , 9burk-PET11 , 9gamm-a0a0d4l7e6 , 9alte-n6vy44 , 9zzzz-a0a0f9x315 , deiml-e8u721 , olean-r4ykl9 , vibga-a0a1z2siq1 , 9burk-a0a0g3bi90 , 9bact-c3ryl0 , 9actn-h6wx58 , idesa-peth , 9bact-g9by57 , acide-PBSA , morsp-lip1 |
| Title : Structural insight into catalytic mechanism of PET hydrolase - Han_2017_Nat.Commun_8_2106 |
| Author(s) : Han X , Liu W , Huang JW , Ma J , Zheng Y , Ko TP , Xu L , Cheng YS , Chen CC , Guo RT |
| Ref : Nat Commun , 8 :2106 , 2017 |
| Abstract : Han_2017_Nat.Commun_8_2106 |
| ESTHER : Han_2017_Nat.Commun_8_2106 |
| PubMedSearch : Han_2017_Nat.Commun_8_2106 |
| PubMedID: 29235460 |
| Gene_locus related to this paper: idesa-peth |
| Title : Tailoring cutinase activity towards polyethylene terephthalate and polyamide 6,6 fibers - Araujo_2007_J.Biotechnol_128_849 |
| Author(s) : Araujo R , Silva C , O'Neill A , Micaelo N , Guebitz G , Soares CMF , Casal M , Cavaco-Paulo A |
| Ref : J Biotechnol , 128 :849 , 2007 |
| Abstract : Araujo_2007_J.Biotechnol_128_849 |
| ESTHER : Araujo_2007_J.Biotechnol_128_849 |
| PubMedSearch : Araujo_2007_J.Biotechnol_128_849 |
| PubMedID: 17306400 |
| Gene_locus related to this paper: fusso-cutas |