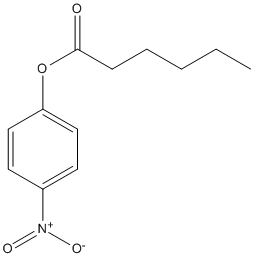
Paranitrophenyl-hexanoate
General
Type : pNP || Chromogen || Water-soluble short chain ester
Chemical_Nomenclature : (4-nitrophenyl) hexanoate
Canonical SMILES : CCCCCC(=O)OC1=CC=C(C=C1)[N+](=O)[O-]
InChI : InChI=1S\/C12H15NO4\/c1-2-3-4-5-12(14)17-11-8-6-10(7-9-11)13(15)16\/h6-9H,2-5H2,1H3
InChIKey : OLRXUEYZKCCEKK-UHFFFAOYSA-N
Other name(s) : pNP-C6, pNPC6, p-NP hexanoate, pNP-hexanoate, 4-Nitrophenyl hexanoate, P-nitrophenyl caproate, Hexanoic Acid 4-Nitrophenyl Ester, D8F, (4-nitrophenyl) hexanoate, AC1LBZXA
References (8)
| Title : Mechanism and Structural Insights Into a Novel Esterase, E53, Isolated From Erythrobacter longus - Ding_2022_Front.Microbiol_12_798194 |
| Author(s) : Ding Y , Nie L , Yang XC , Li Y , Huo YY , Li Z , Gao Y , Cui HL , Li J , Xu XW |
| Ref : Front Microbiol , 12 :798194 , 2022 |
| Abstract : Ding_2022_Front.Microbiol_12_798194 |
| ESTHER : Ding_2022_Front.Microbiol_12_798194 |
| PubMedSearch : Ding_2022_Front.Microbiol_12_798194 |
| PubMedID: 35069500 |
| Gene_locus related to this paper: erylo-E53 |
| Title : The Bacteroidetes Aequorivita sp. and Kaistella jeonii Produce Promiscuous Esterases With PET-Hydrolyzing Activity - Zhang_2022_Front.Microbiol_12_803896 |
| Author(s) : Zhang H , Perez-Garcia P , Dierkes RF , Applegate V , Schumacher J , Chibani CM , Sternagel S , Preuss L , Weigert S , Schmeisser C , Danso D , Pleiss J , Almeida A , Hocker B , Hallam SJ , Schmitz RA , Smits SHJ , Chow J , Streit WR |
| Ref : Front Microbiol , 12 :803896 , 2021 |
| Abstract : Zhang_2022_Front.Microbiol_12_803896 |
| ESTHER : Zhang_2022_Front.Microbiol_12_803896 |
| PubMedSearch : Zhang_2022_Front.Microbiol_12_803896 |
| PubMedID: 35069509 |
| Gene_locus related to this paper: flutr-f2ie04 , 9flao-a0a0c1f4u8 , 9flao-kjj39608 , 9flao-a0a330mq60 |
| Title : Directed evolution of new and improved enzyme functions using an evolutionary intermediate and multidirectional search - Porter_2015_ACS.Chem.Biol_10_611 |
| Author(s) : Porter JL , Boon PL , Murray TP , Huber T , Collyer CA , Ollis DL |
| Ref : ACS Chemical Biology , 10 :611 , 2015 |
| Abstract : Porter_2015_ACS.Chem.Biol_10_611 |
| ESTHER : Porter_2015_ACS.Chem.Biol_10_611 |
| PubMedSearch : Porter_2015_ACS.Chem.Biol_10_611 |
| PubMedID: 25419863 |
| Gene_locus related to this paper: psepu-clcd1 |
| Title : The PE16 (Rv1430) of Mycobacterium tuberculosis Is an Esterase Belonging to Serine Hydrolase Superfamily of Proteins - Sultana_2013_PLoS.One_8_e55320 |
| Author(s) : Sultana R , Vemula MH , Banerjee S , Guruprasad L |
| Ref : PLoS ONE , 8 :e55320 , 2013 |
| Abstract : Sultana_2013_PLoS.One_8_e55320 |
| ESTHER : Sultana_2013_PLoS.One_8_e55320 |
| PubMedSearch : Sultana_2013_PLoS.One_8_e55320 |
| PubMedID: 23383323 |
| Gene_locus related to this paper: myctu-Rv1430 |
| Title : The short form of the recombinant CAL-A-type lipase UM03410 from the smut fungus Ustilago maydis exhibits an inherent trans-fatty acid selectivity - Brundiek_2012_Appl.Microbiol.Biotechnol_94_141 |
| Author(s) : Brundiek H , Sass S , Evitt A , Kourist R , Bornscheuer UT |
| Ref : Applied Microbiology & Biotechnology , 94 :141 , 2012 |
| Abstract : Brundiek_2012_Appl.Microbiol.Biotechnol_94_141 |
| ESTHER : Brundiek_2012_Appl.Microbiol.Biotechnol_94_141 |
| PubMedSearch : Brundiek_2012_Appl.Microbiol.Biotechnol_94_141 |
| PubMedID: 22294433 |
| Gene_locus related to this paper: ustma-q4p903 |
| Title : Molecular cloning and characterization of two thermostable carboxyl esterases from Geobacillus stearothermophilus - Ewis_2004_Gene_329_187 |
| Author(s) : Ewis HE , Abdelal AT , Lu CD |
| Ref : Gene , 329 :187 , 2004 |
| Abstract : Ewis_2004_Gene_329_187 |
| ESTHER : Ewis_2004_Gene_329_187 |
| PubMedSearch : Ewis_2004_Gene_329_187 |
| PubMedID: 15033540 |
| Gene_locus related to this paper: geost-est30 , geost-est50 |
| Title : A novel extracellular esterase from Bacillus subtilis and its conversion to a monoacylglycerol hydrolase - Eggert_2000_Eur.J.Biochem_267_6459 |
| Author(s) : Eggert T , Pencreac'h G , Douchet I , Verger R , Jaeger KE |
| Ref : European Journal of Biochemistry , 267 :6459 , 2000 |
| Abstract : Eggert_2000_Eur.J.Biochem_267_6459 |
| ESTHER : Eggert_2000_Eur.J.Biochem_267_6459 |
| PubMedSearch : Eggert_2000_Eur.J.Biochem_267_6459 |
| PubMedID: 11029590 |
| Gene_locus related to this paper: bacsu-LIPB |
| Title : Purification and partial characterization of a novel thermophilic carboxylesterase with high mesophilic specific activity - Wood_1995_Enzyme.Microb.Technol_17_816 |
| Author(s) : Wood AN , Fernandez-Lafuente R , Cowan DA |
| Ref : Enzyme Microb Technol , 17 :816 , 1995 |
| Abstract : Wood_1995_Enzyme.Microb.Technol_17_816 |
| ESTHER : Wood_1995_Enzyme.Microb.Technol_17_816 |
| PubMedSearch : Wood_1995_Enzyme.Microb.Technol_17_816 |
| PubMedID: 7576531 |
| Gene_locus related to this paper: geost-est50 |