Polycaprolactone
General
Type : Polymer || Polyester || Aliphatic polyester
Chemical_Nomenclature : Poly(hexano-6-lactone)
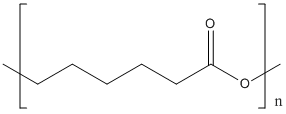
Canonical SMILES :
InChI :
InChIKey :
Other name(s) : PCL, (1,7)-Polyoxepan-2-one, Poly(E''-caprolactone), Poly(E-caprolactone), Poly(sigma-caprolactone), Poly(epsilon-caprolactone), 2-Oxepanone homopolymer, 6-Caprolactone polymer
Target
References (16)
| Title : Characterization of Polymer Degrading Lipases, LIP1 and LIP2 From Pseudomonas chlororaphis PA23 - Mohanan_2022_Front.Bioeng.Biotechnol_10_854298 |
| Author(s) : Mohanan N , Wong CH , Budisa N , Levin DB |
| Ref : Front Bioeng Biotechnol , 10 :854298 , 2022 |
| Abstract : Mohanan_2022_Front.Bioeng.Biotechnol_10_854298 |
| ESTHER : Mohanan_2022_Front.Bioeng.Biotechnol_10_854298 |
| PubMedSearch : Mohanan_2022_Front.Bioeng.Biotechnol_10_854298 |
| PubMedID: 35519608 |
| Gene_locus related to this paper: psecl-LIP1 , psecl-LIP2 |
| Title : Two Extracellular Poly(sigma-caprolactone)-Degrading Enzymes From Pseudomonas hydrolytica sp. DSWY01(T): Purification, Characterization, and Gene Analysis - Li_2022_Front.Bioeng.Biotechnol_10_835847 |
| Author(s) : Li L , Lin X , Bao J , Xia H , Li F |
| Ref : Front Bioeng Biotechnol , 10 :835847 , 2022 |
| Abstract : Li_2022_Front.Bioeng.Biotechnol_10_835847 |
| ESTHER : Li_2022_Front.Bioeng.Biotechnol_10_835847 |
| PubMedSearch : Li_2022_Front.Bioeng.Biotechnol_10_835847 |
| PubMedID: 35372294 |
| Gene_locus related to this paper: psemy-a4xvf4 , psemy-a4y035 |
| Title : The Bacteroidetes Aequorivita sp. and Kaistella jeonii Produce Promiscuous Esterases With PET-Hydrolyzing Activity - Zhang_2022_Front.Microbiol_12_803896 |
| Author(s) : Zhang H , Perez-Garcia P , Dierkes RF , Applegate V , Schumacher J , Chibani CM , Sternagel S , Preuss L , Weigert S , Schmeisser C , Danso D , Pleiss J , Almeida A , Hocker B , Hallam SJ , Schmitz RA , Smits SHJ , Chow J , Streit WR |
| Ref : Front Microbiol , 12 :803896 , 2021 |
| Abstract : Zhang_2022_Front.Microbiol_12_803896 |
| ESTHER : Zhang_2022_Front.Microbiol_12_803896 |
| PubMedSearch : Zhang_2022_Front.Microbiol_12_803896 |
| PubMedID: 35069509 |
| Gene_locus related to this paper: flutr-f2ie04 , 9flao-a0a0c1f4u8 , 9flao-kjj39608 , 9flao-a0a330mq60 |
| Title : An extracellular lipase from Amycolatopsis mediterannei is a cutinase with plastic degrading activity - Tan_2021_Comput.Struct.Biotechnol.J_19_869 |
| Author(s) : Tan Y , Henehan GT , Kinsella GK , Ryan BJ |
| Ref : Comput Struct Biotechnol J , 19 :869 , 2021 |
| Abstract : Tan_2021_Comput.Struct.Biotechnol.J_19_869 |
| ESTHER : Tan_2021_Comput.Struct.Biotechnol.J_19_869 |
| PubMedSearch : Tan_2021_Comput.Struct.Biotechnol.J_19_869 |
| PubMedID: 33598102 |
| Gene_locus related to this paper: amymu-a0a0h3des9 |
| Title : Penicillium fellutanum lipase as a green and ecofriendly biocatalyst for depolymerization of poly (sigma-caprolactone): Biochemical, kinetic, and thermodynamic investigations - Amin_2021_Biotechnol.Appl.Biochem__ |
| Author(s) : Amin M , Bhatti HN , Nawaz S , Bilal M |
| Ref : Biotechnol Appl Biochem , : , 2021 |
| Abstract : Amin_2021_Biotechnol.Appl.Biochem__ |
| ESTHER : Amin_2021_Biotechnol.Appl.Biochem__ |
| PubMedSearch : Amin_2021_Biotechnol.Appl.Biochem__ |
| PubMedID: 33559904 |
| Title : Biodegradation mechanism of polycaprolactone by a novel esterase MGS0156: a QM\/MM approach - Feng_2020_Environ.Sci.Process.Impacts_22_2332 |
| Author(s) : Feng S , Yue Y , Chen J , Zhou J , Li Y , Zhang Q |
| Ref : Environ Sci Process Impacts , 22 :2332 , 2020 |
| Abstract : Feng_2020_Environ.Sci.Process.Impacts_22_2332 |
| ESTHER : Feng_2020_Environ.Sci.Process.Impacts_22_2332 |
| PubMedSearch : Feng_2020_Environ.Sci.Process.Impacts_22_2332 |
| PubMedID: 33146659 |
| Gene_locus related to this paper: 9zzzz-A0A0G3FEJ8 |
| Title : In silico Screening and Heterologous Expression of a Polyethylene Terephthalate Hydrolase (PETase)-Like Enzyme (SM14est) With Polycaprolactone (PCL)-Degrading Activity, From the Marine Sponge-Derived Strain Streptomyces sp. SM14 - Almeida_2019_Front.Microbiol_10_2187 |
| Author(s) : Almeida EL , Rincon AFC , Jackson SA , Dobson ADW , Carrillo Rincon AF |
| Ref : Front Microbiol , 10 :2187 , 2019 |
| Abstract : Almeida_2019_Front.Microbiol_10_2187 |
| ESTHER : Almeida_2019_Front.Microbiol_10_2187 |
| PubMedSearch : Almeida_2019_Front.Microbiol_10_2187 |
| PubMedID: 31632361 |
| Gene_locus related to this paper: 9actn-SM14est |
| Title : Screening and Characterization of Novel Polyesterases from Environmental Metagenomes with High Hydrolytic Activity against Synthetic Polyesters - Hajighasemi_2018_Environ.Sci.Technol_52_12388 |
| Author(s) : Hajighasemi M , Tchigvintsev A , Nocek B , Flick R , Popovic A , Hai T , Khusnutdinova AN , Brown G , Xu X , Cui H , Anstett J , Chernikova TN , Bruls T , Le Paslier D , Yakimov MM , Joachimiak A , Golyshina OV , Savchenko A , Golyshin PN , Edwards EA , Yakunin AF |
| Ref : Environ Sci Technol , 52 :12388 , 2018 |
| Abstract : Hajighasemi_2018_Environ.Sci.Technol_52_12388 |
| ESTHER : Hajighasemi_2018_Environ.Sci.Technol_52_12388 |
| PubMedSearch : Hajighasemi_2018_Environ.Sci.Technol_52_12388 |
| PubMedID: 30284819 |
| Gene_locus related to this paper: 9zzzz-a0a0g3fj39 , 9zzzz-a0a0g3fj48 , 9zzzz-A0A0G3FEJ8 , 9bact-a4uz10 |
| Title : Functional characterization and structural modeling of synthetic polyester-degrading hydrolases from Thermomonospora curvata - Wei_2014_AMB.Express_4_44 |
| Author(s) : Wei R , Oeser T , Then J , Kuhn N , Barth M , Schmidt J , Zimmermann W |
| Ref : AMB Express , 4 :44 , 2014 |
| Abstract : Wei_2014_AMB.Express_4_44 |
| ESTHER : Wei_2014_AMB.Express_4_44 |
| PubMedSearch : Wei_2014_AMB.Express_4_44 |
| PubMedID: 25405080 |
| Gene_locus related to this paper: thecd-d1a9g5 , thecd-d1a2h1 |
| Title : Identification and comparison of cutinases for synthetic polyester degradation - Baker_2012_Appl.Microbiol.Biotechnol_93_229 |
| Author(s) : Baker PJ , Poultney C , Liu Z , Gross R , Montclare JK |
| Ref : Applied Microbiology & Biotechnology , 93 :229 , 2012 |
| Abstract : Baker_2012_Appl.Microbiol.Biotechnol_93_229 |
| ESTHER : Baker_2012_Appl.Microbiol.Biotechnol_93_229 |
| PubMedSearch : Baker_2012_Appl.Microbiol.Biotechnol_93_229 |
| PubMedID: 21713515 |
| Gene_locus related to this paper: humin-cut |
| Title : Isolation of a novel cutinase homolog with polyethylene terephthalate-degrading activity from leaf-branch compost by using a metagenomic approach - Sulaiman_2012_Appl.Environ.Microbiol_78_1556 |
| Author(s) : Sulaiman S , Yamato S , Kanaya E , Kim JJ , Koga Y , Takano K , Kanaya S |
| Ref : Applied Environmental Microbiology , 78 :1556 , 2012 |
| Abstract : Sulaiman_2012_Appl.Environ.Microbiol_78_1556 |
| ESTHER : Sulaiman_2012_Appl.Environ.Microbiol_78_1556 |
| PubMedSearch : Sulaiman_2012_Appl.Environ.Microbiol_78_1556 |
| PubMedID: 22194294 |
| Gene_locus related to this paper: 9bact-g9by57 |
| Title : Cutinolytic esterase activity of bacteria isolated from mixed-plant compost and characterization of a cutinase gene from Pseudomonas pseudoalcaligenes - Inglis_2011_Can.J.Microbiol_57_902 |
| Author(s) : Inglis GD , Yanke LJ , Selinger LB |
| Ref : Can J Microbiol , 57 :902 , 2011 |
| Abstract : Inglis_2011_Can.J.Microbiol_57_902 |
| ESTHER : Inglis_2011_Can.J.Microbiol_57_902 |
| PubMedSearch : Inglis_2011_Can.J.Microbiol_57_902 |
| PubMedID: 22029433 |
| Gene_locus related to this paper: pseol-e9kjl1 |
| Title : Enzymatic surface hydrolysis of poly(ethylene terephthalate) and bis(benzoyloxyethyl) terephthalate by lipase and cutinase in the presence of surface active molecules - Eberl_2009_J.Biotechnol_143_207 |
| Author(s) : Eberl A , Heumann S , Bruckner T , Araujo R , Cavaco-Paulo A , Kaufmann F , Kroutil W , Guebitz GM |
| Ref : J Biotechnol , 143 :207 , 2009 |
| Abstract : Eberl_2009_J.Biotechnol_143_207 |
| ESTHER : Eberl_2009_J.Biotechnol_143_207 |
| PubMedSearch : Eberl_2009_J.Biotechnol_143_207 |
| PubMedID: 19616594 |
| Title : Structural and functional studies of Aspergillus oryzae cutinase: enhanced thermostability and hydrolytic activity of synthetic ester and polyester degradation - Liu_2009_J.Am.Chem.Soc_131_15711 |
| Author(s) : Liu Z , Gosser Y , Baker PJ , Ravee Y , Lu Z , Alemu G , Li H , Butterfoss GL , Kong XP , Gross R , Montclare JK |
| Ref : Journal of the American Chemical Society , 131 :15711 , 2009 |
| Abstract : Liu_2009_J.Am.Chem.Soc_131_15711 |
| ESTHER : Liu_2009_J.Am.Chem.Soc_131_15711 |
| PubMedSearch : Liu_2009_J.Am.Chem.Soc_131_15711 |
| PubMedID: 19810726 |
| Gene_locus related to this paper: aspor-cutas |
| Title : Cutinase-like enzyme from the yeast Cryptococcus sp. strain S-2 hydrolyzes polylactic acid and other biodegradable plastics - Masaki_2005_Appl.Environ.Microbiol_71_7548 |
| Author(s) : Masaki K , Kamini NR , Ikeda H , Iefuji H |
| Ref : Applied Environmental Microbiology , 71 :7548 , 2005 |
| Abstract : Masaki_2005_Appl.Environ.Microbiol_71_7548 |
| ESTHER : Masaki_2005_Appl.Environ.Microbiol_71_7548 |
| PubMedSearch : Masaki_2005_Appl.Environ.Microbiol_71_7548 |
| PubMedID: 16269800 |
| Gene_locus related to this paper: crysp-Q874E9 |
| Title : Fusarium polycaprolactone depolymerase is cutinase - Murphy_1996_Appl.Environ.Microbiol_62_456 |
| Author(s) : Murphy CA , Cameron JA , Huang SJ , Vinopal RT |
| Ref : Applied Environmental Microbiology , 62 :456 , 1996 |
| Abstract : Murphy_1996_Appl.Environ.Microbiol_62_456 |
| ESTHER : Murphy_1996_Appl.Environ.Microbiol_62_456 |
| PubMedSearch : Murphy_1996_Appl.Environ.Microbiol_62_456 |
| PubMedID: 8593048 |
| Gene_locus related to this paper: fusso-CUT2 , fusso-CUT3 |