trans-Bifenthrin
General
Type : pyrethroid || Insecticide || Trifluoro || Not A\/B H target || Carbamate || Cyclopropane
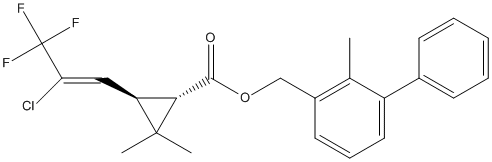
Chemical_Nomenclature : (2-methyl-3-phenylphenyl)methyl (1S,3R)-3-[(Z)-2-chloro-3,3,3-trifluoroprop-1-enyl]-2,2-dimethylcyclopropane-1-carboxylate
Canonical SMILES : CC1=C(C=CC=C1COC(=O)C2C(C2(C)C)C=C(C(F)(F)F)Cl)C3=CC=CC=C3
InChI : InChI=1S\/C23H22ClF3O2\/c1-14-16(10-7-11-17(14)15-8-5-4-6-9-15)13-29-21(28)20-18(22(20,2)3)12-19(24)23(25,26)27\/h4-12,18,20H,13H2,1-3H3\/b19-12-\/t18-,20+\/m0\/s1
InChIKey : OMFRMAHOUUJSGP-GUGVIDNXSA-N
Other name(s) : Trans-Bifenthrin, Bifenthrin, trans, DTXSID1058501, ZINC1532089, CJ-24130, Trans-Bifenthtin, 8NL
Target
Families : HNLyase_Bact, Carb_B_Arthropoda
References (2)
| Title : Characterization of the insecticide detoxification carboxylesterase Boest1 from Bradysia odoriphaga Yang et Zhang (Diptera:Sciaridae) - Ding_2021_Pest.Manag.Sci__ |
| Author(s) : Ding Q , Xu X , Sang Z , Wang R , Ullah F , Gao X , Song D |
| Ref : Pest Manag Sci , : , 2021 |
| Abstract : Ding_2021_Pest.Manag.Sci__ |
| ESTHER : Ding_2021_Pest.Manag.Sci__ |
| PubMedSearch : Ding_2021_Pest.Manag.Sci__ |
| PubMedID: 34596943 |
| Gene_locus related to this paper: 9dipt-Boest1 , braco-est1 |
| Title : Pyrethroid Carboxylesterase PytH from Sphingobium faniae JZ-2: Structure and Catalytic Mechanism - Xu_2020_Appl.Environ.Microbiol_86_e02971 |
| Author(s) : Xu D , Gao Y , Sun B , Ran T , Zeng L , He J , Wang W |
| Ref : Applied Environmental Microbiology , 86 :e02971 , 2020 |
| Abstract : Xu_2020_Appl.Environ.Microbiol_86_e02971 |
| ESTHER : Xu_2020_Appl.Environ.Microbiol_86_e02971 |
| PubMedSearch : Xu_2020_Appl.Environ.Microbiol_86_e02971 |
| PubMedID: 32303545 |
| Gene_locus related to this paper: sphwj-c0la90 |