Phenmedipham
Substrate of artox-phhyd. Herbicide Manufacturers: KVK Agro, AgrEvo, Esbjerg Kemi. Also inhibitor of fatty acid amide hydrolase (FAAH) (not alpha/beta hydrolase) Vincent et al. 2009 Bioorg Med Chem Lett. 2009 Dec 1;19(23):6793-6 PMID:19850474
General
Type : Carbamate,Herbicide,Not A\/B H target,FAAH inhibitor
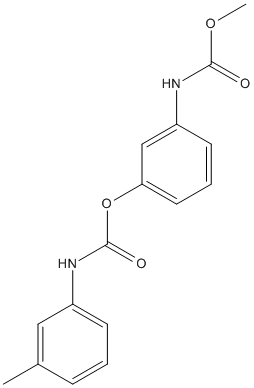
Chemical_Nomenclature : [3-(methoxycarbonylamino)phenyl] N-(3-methylphenyl)carbamate
Canonical SMILES : CC1=CC(=CC=C1)NC(=O)OC2=CC=CC(=C2)NC(=O)OC
InChI : InChI=1S\/C16H16N2O4\/c1-11-5-3-6-12(9-11)18-16(20)22-14-8-4-7-13(10-14)17-15(19)21-2\/h3-10H,1-2H3,(H,17,19)(H,18,20)
InChIKey : IDOWTHOLJBTAFI-UHFFFAOYSA-N
Other name(s) : Herbasan,Betanal,Betasana,3-((Methoxycarbonyl)amino)phenyl (3-methylphenyl)carbamate,3-(Carbomethoxyamino)phenyl 3-methylcarbanilate,3-(Methylphenyl)carbamic acid 3-((methoxycarbonyl)amino)phenyl ester,3-Methoxycarbonylaminophenyl N-3'-methylphenylcarbamate
MW : 300.31
Formula : C16H16N2O4
CAS_number : 13684-63-4
PubChem : 24744
UniChem : IDOWTHOLJBTAFI-UHFFFAOYSA-N
IUPHAR :
Wikipedia :
Target
References (4)
| Title : Biochemical characterization of cholinesterases in Enchytraeus albidus and assessment of in vivo and in vitro effects of different soil properties, copper and phenmedipham - Howcroft_2011_Ecotoxicology_20_119 |
| Author(s) : Howcroft CF , Gravato C , Amorim MJ , Novais SC , Soares AM , Guilhermino L |
| Ref : Ecotoxicology , 20 :119 , 2011 |
| Abstract : Howcroft_2011_Ecotoxicology_20_119 |
| ESTHER : Howcroft_2011_Ecotoxicology_20_119 |
| PubMedSearch : Howcroft_2011_Ecotoxicology_20_119 |
| PubMedID: 21080225 |
| Title : Biological monitoring of phenmedipham: determination of m-toluidine in urine - Schettgen_2001_Arch.Toxicol_75_145 |
| Author(s) : Schettgen T , Weiss T , Angerer J |
| Ref : Archives of Toxicology , 75 :145 , 2001 |
| Abstract : Schettgen_2001_Arch.Toxicol_75_145 |
| ESTHER : Schettgen_2001_Arch.Toxicol_75_145 |
| PubMedSearch : Schettgen_2001_Arch.Toxicol_75_145 |
| PubMedID: 11409536 |
| Title : Purification and properties of an Arthrobacter oxydans P52 carbamate hydrolase specific for the herbicide phenmedipham and nucleotide sequence of the corresponding gene - Pohlenz_1992_J.Bacteriol_174_6600 |
| Author(s) : Pohlenz HD , Boidol W , Schuttke I , Streber WR |
| Ref : Journal of Bacteriology , 174 :6600 , 1992 |
| Abstract : Pohlenz_1992_J.Bacteriol_174_6600 |
| ESTHER : Pohlenz_1992_J.Bacteriol_174_6600 |
| PubMedSearch : Pohlenz_1992_J.Bacteriol_174_6600 |
| PubMedID: 1400211 |
| Gene_locus related to this paper: artox-phhyd |
| Title : [Radiometric determination of 11 carbamate pesticides in the nanogram and subnanongram ranges by means of cholinesterase inhibition] - Schmid_1977_Nahrung_21_311 |
| Author(s) : Schmid ER , Damboritz W , Markl P |
| Ref : Nahrung , 21 :311 , 1977 |
| Abstract : Schmid_1977_Nahrung_21_311 |
| ESTHER : Schmid_1977_Nahrung_21_311 |
| PubMedSearch : Schmid_1977_Nahrung_21_311 |
| PubMedID: 406567 |