Paranitrobutyranilide
General
Type : Amidase substrate, pNP, Chromogen
Chemical_Nomenclature :
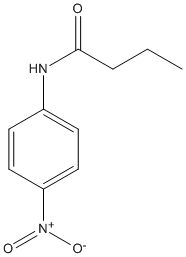
Canonical SMILES : CCCC(=O)NC1=CC=C(C=C1)[N+](=O)[O-]
InChI : InChI=1S\/C10H12N2O3\/c1-2-3-10(13)11-8-4-6-9(7-5-8)12(14)15\/h4-7H,2-3H2,1H3,(H,11,13)
InChIKey : YATAVUWVWRFIRI-UHFFFAOYSA-N
Other name(s) : N-(4-nitrophenyl)butanamide || p-nitrobutyranilide || NSC404442 || Oprea1_433813
MW : 208.21
Formula : C10H12N2O3
CAS_number : 54191-12-7
PubChem : 346259
UniChem : YATAVUWVWRFIRI-UHFFFAOYSA-N
Target
Structures : No structure
Families : Cutinase, Canar_LipB, Carb_B_Bacteria
References (6)
| Title : Cutinase-Catalyzed Polyester-Polyurethane Degradation: Elucidation of the Hydrolysis Mechanism - Di Bisceglie_2022_Polymers.(Basel)_14_411 |
| Author(s) : Di Bisceglie F , Quartinello F , Vielnascher R , Guebitz GM , Pellis A |
| Ref : Polymers (Basel) , 14 :411 , 2022 |
| Abstract : |
| PubMedSearch : Di Bisceglie_2022_Polymers.(Basel)_14_411 |
| PubMedID: 35160402 |
| Gene_locus related to this paper: humin-cut |
| Title : Cutinase-Catalyzed Polyester-Polyurethane Degradation: Elucidation of the Hydrolysis Mechanism - Di Bisceglie_2022_Polymers.(Basel)_14_411 |
| Author(s) : Di Bisceglie F , Quartinello F , Vielnascher R , Guebitz GM , Pellis A |
| Ref : Polymers (Basel) , 14 :411 , 2022 |
| Abstract : |
| PubMedSearch : Di Bisceglie_2022_Polymers.(Basel)_14_411 |
| PubMedID: 35160402 |
| Gene_locus related to this paper: humin-cut |
| Title : Cutinase-Catalyzed Polyester-Polyurethane Degradation: Elucidation of the Hydrolysis Mechanism - Di Bisceglie_2022_Polymers.(Basel)_14_411 |
| Author(s) : Di Bisceglie F , Quartinello F , Vielnascher R , Guebitz GM , Pellis A |
| Ref : Polymers (Basel) , 14 :411 , 2022 |
| Abstract : |
| PubMedSearch : Di Bisceglie_2022_Polymers.(Basel)_14_411 |
| PubMedID: 35160402 |
| Gene_locus related to this paper: humin-cut |
| Title : Understanding promiscuous amidase activity of an esterase from Bacillus subtilis - |
| Author(s) : Kourist R , Bartsch S , Fransson L , Hult K , Bornscheuer UT |
| Ref : Chembiochem , 9 :67 , 2008 |
| PubMedID: 18022973 |
| Gene_locus related to this paper: bacsu-pnbae , canar-LipB |
| Title : Understanding promiscuous amidase activity of an esterase from Bacillus subtilis - |
| Author(s) : Kourist R , Bartsch S , Fransson L , Hult K , Bornscheuer UT |
| Ref : Chembiochem , 9 :67 , 2008 |
| PubMedID: 18022973 |
| Gene_locus related to this paper: bacsu-pnbae , canar-LipB |
| Title : Understanding promiscuous amidase activity of an esterase from Bacillus subtilis - |
| Author(s) : Kourist R , Bartsch S , Fransson L , Hult K , Bornscheuer UT |
| Ref : Chembiochem , 9 :67 , 2008 |
| PubMedID: 18022973 |
| Gene_locus related to this paper: bacsu-pnbae , canar-LipB |