Dienelactone
General
Type : Lactone
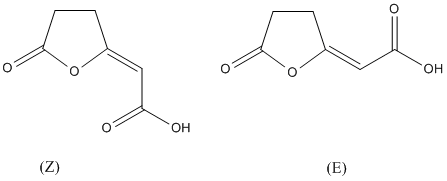
Chemical_Nomenclature : (2Z)-2-(5-oxofuran-2-ylidene)acetic acid
Canonical SMILES : C1=CC(=O)OC1=CC(=O)O
InChI : InChI=1S\/C6H4O4\/c7-5(8)3-4-1-2-6(9)10-4\/h1-3H,(H,7,8)\/b4-3-
InChIKey : AYFXPGXAZMFWNH-ONEGZZNKSA-N || AYFXPGXAZMFWNH-ARJAWSKDSA-N
Other name(s) : (5-oxofuran-2(5H)-ylidene)acetic acid, Cis-4-carboxymethylenebut-2-en-4-olide, (5-Oxo-2(5H)-furanylidene)acetic acid, 4-carboxymethylenebut-2-en-4-olide, (2E)-(5-oxofuran-2(5H)-ylidene)acetic acid, Trans-4-Carboxymethylenebut-2-en-4-olide, AC1NQXEN, SCHEMBL891606, CHEBI:38107
Target
Families : Dienelactone_hydrolase
References (8)
| Title : Biochemical and structural characterization of a novel cold-active esterase-like protein from the psychrophilic yeast Glaciozyma antarctica - Hashim_2018_Extremophiles_22_607 |
| Author(s) : Hashim NHF , Mahadi NM , Illias RM , Feroz SR , Abu Bakar FD , Murad AMA |
| Ref : Extremophiles , 22 :607 , 2018 |
| Abstract : Hashim_2018_Extremophiles_22_607 |
| ESTHER : Hashim_2018_Extremophiles_22_607 |
| PubMedSearch : Hashim_2018_Extremophiles_22_607 |
| PubMedID: 29556723 |
| Gene_locus related to this paper: 9basi-a0a221se81 |
| Title : Two structurally different dienelactone hydrolases (TfdEI and TfdEII) from Cupriavidus necator JMP134 plasmid pJP4 catalyse cis- and trans-dienelactones with similar efficiency - Kumar_2014_PLoS.One_9_e101801 |
| Author(s) : Kumar A , Pillay B , Olaniran AO |
| Ref : PLoS ONE , 9 :e101801 , 2014 |
| Abstract : Kumar_2014_PLoS.One_9_e101801 |
| ESTHER : Kumar_2014_PLoS.One_9_e101801 |
| PubMedSearch : Kumar_2014_PLoS.One_9_e101801 |
| PubMedID: 25054964 |
| Gene_locus related to this paper: alceu-tfe1 , alceu-tfe2 |
| Title : Substrate-induced conformational change and isomerase activity of dienelactone hydrolase and its site-specific mutants - Walker_2012_Chembiochem_13_1645 |
| Author(s) : Walker I , Hennessy JE , Ollis DL , Easton CJ |
| Ref : Chembiochem , 13 :1645 , 2012 |
| Abstract : Walker_2012_Chembiochem_13_1645 |
| ESTHER : Walker_2012_Chembiochem_13_1645 |
| PubMedSearch : Walker_2012_Chembiochem_13_1645 |
| PubMedID: 22761053 |
| Title : Monitoring key reactions in degradation of chloroaromatics by in situ (1)H nuclear magnetic resonance: solution structures of metabolites formed from cis-dienelactone - Pieper_2002_J.Bacteriol_184_1466 |
| Author(s) : Pieper DH , Pollmann K , Nikodem P , Gonzalez B , Wray V |
| Ref : Journal of Bacteriology , 184 :1466 , 2002 |
| Abstract : Pieper_2002_J.Bacteriol_184_1466 |
| ESTHER : Pieper_2002_J.Bacteriol_184_1466 |
| PubMedSearch : Pieper_2002_J.Bacteriol_184_1466 |
| PubMedID: 11844781 |
| Title : Substrate-induced activation of dienelactone hydrolase: an enzyme with a naturally occurring Cys-His-Asp triad - Cheah_1993_Protein.Eng_6_575 |
| Author(s) : Cheah E , Austin C , Ashley GW , Ollis D |
| Ref : Protein Engineering , 6 :575 , 1993 |
| Abstract : Cheah_1993_Protein.Eng_6_575 |
| ESTHER : Cheah_1993_Protein.Eng_6_575 |
| PubMedSearch : Cheah_1993_Protein.Eng_6_575 |
| PubMedID: 8234228 |
| Gene_locus related to this paper: psepu-clcd1 |
| Title : Catalysis by dienelactone hydrolase: a variation on the protease mechanism - Cheah_1993_Proteins_16_64 |
| Author(s) : Cheah E , Ashley GW , Gary J , Ollis D |
| Ref : Proteins , 16 :64 , 1993 |
| Abstract : Cheah_1993_Proteins_16_64 |
| ESTHER : Cheah_1993_Proteins_16_64 |
| PubMedSearch : Cheah_1993_Proteins_16_64 |
| PubMedID: 8497485 |
| Gene_locus related to this paper: psepu-clcd1 |
| Title : Refined structure of dienelactone hydrolase at 1.8 A - Pathak_1990_J.Mol.Biol_214_497 |
| Author(s) : Pathak D , Ollis D |
| Ref : Journal of Molecular Biology , 214 :497 , 1990 |
| Abstract : Pathak_1990_J.Mol.Biol_214_497 |
| ESTHER : Pathak_1990_J.Mol.Biol_214_497 |
| PubMedSearch : Pathak_1990_J.Mol.Biol_214_497 |
| PubMedID: 2380986 |
| Gene_locus related to this paper: psepu-clcd1 |
| Title : X-ray crystallographic structure of dienelactone hydrolase at 2.8 A - Pathak_1988_J.Mol.Biol_204_435 |
| Author(s) : Pathak D , Ngai KL , Ollis D |
| Ref : Journal of Molecular Biology , 204 :435 , 1988 |
| Abstract : Pathak_1988_J.Mol.Biol_204_435 |
| ESTHER : Pathak_1988_J.Mol.Biol_204_435 |
| PubMedSearch : Pathak_1988_J.Mol.Biol_204_435 |
| PubMedID: 3221394 |
| Gene_locus related to this paper: psepu-clcd1 |