Methyl-Gallate
General
Type : Derivative of Gallate || Benzoate
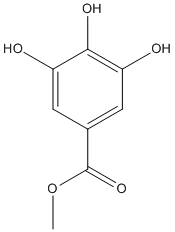
Chemical_Nomenclature : methyl 3,4,5-trihydroxybenzoate
Canonical SMILES : COC(=O)C1=CC(=C(C(=C1)O)O)O
InChI : InChI=1S\/C8H8O5\/c1-13-8(12)4-2-5(9)7(11)6(10)3-4\/h2-3,9-11H,1H3
InChIKey : FBSFWRHWHYMIOG-UHFFFAOYSA-N
Other name(s) : Methyl Gallate, Methyl 3,4,5-trihydroxybenzoate, CHEMBL65675, SCHEMBL39513, ZINC21789, CHEBI:145828, Gallic acid methyl ester, Methylgallate
Target
Families : Tannase_Bact, Tannase, Plant_carboxylesterase
References (9)
| Title : Discovery and characterization of tannase genes in plants: roles in hydrolysis of tannins - Dai_2020_New.Phytol_226_1104 |
| Author(s) : Dai X , Liu Y , Zhuang J , Yao S , Liu L , Jiang X , Zhou K , Wang Y , Xie D , Bennetzen JL , Gao L , Xia T |
| Ref : New Phytol , 226 :1104 , 2020 |
| Abstract : Dai_2020_New.Phytol_226_1104 |
| ESTHER : Dai_2020_New.Phytol_226_1104 |
| PubMedSearch : Dai_2020_New.Phytol_226_1104 |
| PubMedID: 32061142 |
| Gene_locus related to this paper: camsi-a0a5b9g8c2 , jugre-JrTA , fraan-FaTA , vitvi-VvTA , citcl-CcTA , dioka-DkTA , camsi-CsTA |
| Title : Mutational analysis of Kex2 recognition sites and a disulfide bond in tannase from Aspergillus oryzae - Koseki_2017_Biochem.Biophys.Res.Commun_482_1165 |
| Author(s) : Koseki T , Otsuka M , Mizuno T , Shiono Y |
| Ref : Biochemical & Biophysical Research Communications , 482 :1165 , 2017 |
| Abstract : Koseki_2017_Biochem.Biophys.Res.Commun_482_1165 |
| ESTHER : Koseki_2017_Biochem.Biophys.Res.Commun_482_1165 |
| PubMedSearch : Koseki_2017_Biochem.Biophys.Res.Commun_482_1165 |
| PubMedID: 27919681 |
| Gene_locus related to this paper: aspor-tan |
| Title : Gallic acid-based alkyl esters synthesis in a water-free system by celite-bound lipase of Bacillus licheniformis SCD11501 - Sharma_2015_Biotechnol.Prog_31_715 |
| Author(s) : Sharma S , Kanwar SS , Dogra P , Chauhan GS |
| Ref : Biotechnol Prog , 31 :715 , 2015 |
| Abstract : Sharma_2015_Biotechnol.Prog_31_715 |
| ESTHER : Sharma_2015_Biotechnol.Prog_31_715 |
| PubMedSearch : Sharma_2015_Biotechnol.Prog_31_715 |
| PubMedID: 25737230 |
| Title : A Lactobacillus plantarum Esterase Active on a Broad Range of Phenolic Esters - Esteban-Torres_2015_Appl.Environ.Microbiol_81_3235 |
| Author(s) : Esteban-Torres M , Landete JM , Reveron I , Santamaria L , de Las Rivas B , Munoz R |
| Ref : Applied Environmental Microbiology , 81 :3235 , 2015 |
| Abstract : Esteban-Torres_2015_Appl.Environ.Microbiol_81_3235 |
| ESTHER : Esteban-Torres_2015_Appl.Environ.Microbiol_81_3235 |
| PubMedSearch : Esteban-Torres_2015_Appl.Environ.Microbiol_81_3235 |
| PubMedID: 25746986 |
| Gene_locus related to this paper: lacpl-Est.1092 |
| Title : Tannin degradation by a novel tannase enzyme present in some Lactobacillus plantarum strains - Jimenez_2014_Appl.Environ.Microbiol_80_2991 |
| Author(s) : Jimenez N , Esteban-Torres M , Mancheno JM , de Las Rivas B , Munoz R |
| Ref : Applied Environmental Microbiology , 80 :2991 , 2014 |
| Abstract : Jimenez_2014_Appl.Environ.Microbiol_80_2991 |
| ESTHER : Jimenez_2014_Appl.Environ.Microbiol_80_2991 |
| PubMedSearch : Jimenez_2014_Appl.Environ.Microbiol_80_2991 |
| PubMedID: 24610854 |
| Gene_locus related to this paper: lacpn-d7vbf4 |
| Title : Catalytical Properties of Free and Immobilized Aspergillus niger Tannase - Flores-Maltos_2011_Enzyme.Res_2011_768183 |
| Author(s) : Flores-Maltos A , Rodriguez-Duran LV , Renovato J , Contreras JC , Rodriguez R , Aguilar CN |
| Ref : Enzyme Res , 2011 :768183 , 2011 |
| Abstract : Flores-Maltos_2011_Enzyme.Res_2011_768183 |
| ESTHER : Flores-Maltos_2011_Enzyme.Res_2011_768183 |
| PubMedSearch : Flores-Maltos_2011_Enzyme.Res_2011_768183 |
| PubMedID: 21918717 |
| Title : Acidophilic tannase from marine Aspergillus awamori BTMFW032 - Beena_2010_J.Microbiol.Biotechnol_20_1403 |
| Author(s) : Beena PS , Soorej MB , Elyas KK , Sarita GB , Chandrasekaran M |
| Ref : J Microbiol Biotechnol , 20 :1403 , 2010 |
| Abstract : Beena_2010_J.Microbiol.Biotechnol_20_1403 |
| ESTHER : Beena_2010_J.Microbiol.Biotechnol_20_1403 |
| PubMedSearch : Beena_2010_J.Microbiol.Biotechnol_20_1403 |
| PubMedID: 21030825 |
| Title : Purification and characterization of two cold-adapted extracellular tannin acyl hydrolases from an Antarctic strain Verticillium sp. P9 - Kasieczka-Burnecka_2007_Appl.Microbiol.Biotechnol_77_77 |
| Author(s) : Kasieczka-Burnecka M , Kuc K , Kalinowska H , Knap M , Turkiewicz M |
| Ref : Applied Microbiology & Biotechnology , 77 :77 , 2007 |
| Abstract : Kasieczka-Burnecka_2007_Appl.Microbiol.Biotechnol_77_77 |
| ESTHER : Kasieczka-Burnecka_2007_Appl.Microbiol.Biotechnol_77_77 |
| PubMedSearch : Kasieczka-Burnecka_2007_Appl.Microbiol.Biotechnol_77_77 |
| PubMedID: 17786433 |
| Title : Visual reading method for detection of bacterial tannase - Osawa_1993_Appl.Environ.Microbiol_59_1251 |
| Author(s) : Osawa R , Walsh TP |
| Ref : Applied Environmental Microbiology , 59 :1251 , 1993 |
| Abstract : Osawa_1993_Appl.Environ.Microbiol_59_1251 |
| ESTHER : Osawa_1993_Appl.Environ.Microbiol_59_1251 |
| PubMedSearch : Osawa_1993_Appl.Environ.Microbiol_59_1251 |
| PubMedID: 16348918 |