3HB-CoA
General
Type : CoA || Aminopurin
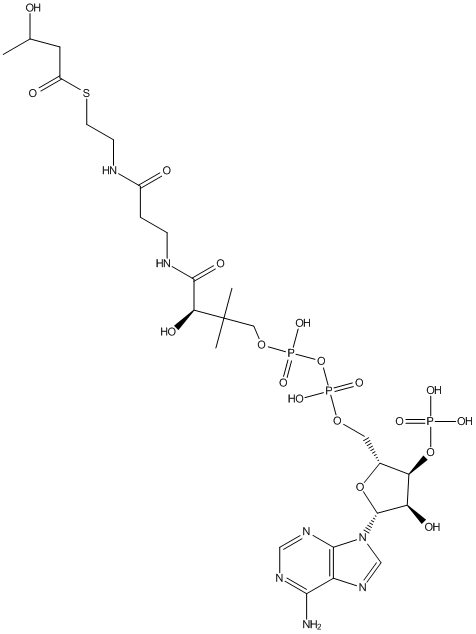
Chemical_Nomenclature : S-[2-[3-[[(2R)-4-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-hydroxy-3-phosphonooxyoxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-hydroxyphosphoryl]oxy-2-hydroxy-3,3-dimethylbutanoyl]amino]propanoylamino]ethyl] 3-hydroxybutanethioate
Canonical SMILES : CC(CC(=O)SCCNC(=O)CCNC(=O)C(C(C)(C)COP(=O)(O)OP(=O)(O)OCC1C(C(C(O1)N2C=NC3=C(N=CN=C32)N)O)OP(=O)(O)O)O)O
InChI : InChI=1S\/C25H42N7O18P3S\/c1-13(33)8-16(35)54-7-6-27-15(34)4-5-28-23(38)20(37)25(2,3)10-47-53(44,45)50-52(42,43)46-9-14-19(49-51(39,40)41)18(36)24(48-14)32-12-31-17-21(26)29-11-30-22(17)32\/h11-14,18-20,24,33,36-37H,4-10H2,1-3H3,(H,27,34)(H,28,38)(H,42,43)(H,44,45)(H2,26,29,30)(H2,39,40,41)\/t13?,14-,18-,19-,20+,24-\/m1\/s1
InChIKey : QHHKKMYHDBRONY-RMNRSTNRSA-N
Other name(s) : (R)-3-hydroxybutyryl-CoA, CHEBI:57315, (3R)-3-hydroxybutanoyl-CoA, 3-hydroxybutanoyl-CoA, 3-hydroxybutyryl-CoA, beta-hydroxybutyryl-CoA, 3-Hydroxybutyryl-coenzyme A, beta-Hydroxybutyryl-coenzyme A, SCHEMBL60564, CHEBI:37050, DB03612
Target
Families : PHA_synth_I, PHA_synth_III_C
References (6)
| Title : Polyester synthases: natural catalysts for plastics - Rehm_2003_Biochem.J_376_15 |
| Author(s) : Rehm BH |
| Ref : Biochemical Journal , 376 :15 , 2003 |
| Abstract : Rehm_2003_Biochem.J_376_15 |
| ESTHER : Rehm_2003_Biochem.J_376_15 |
| PubMedSearch : Rehm_2003_Biochem.J_376_15 |
| PubMedID: 12954080 |
| Title : In vitro evolution of a polyhydroxybutyrate synthase by intragenic suppression-type mutagenesis - Taguchi_2002_J.Biochem_131_801 |
| Author(s) : Taguchi S , Nakamura H , Hiraishi T , Yamato I , Doi Y |
| Ref : J Biochem , 131 :801 , 2002 |
| Abstract : Taguchi_2002_J.Biochem_131_801 |
| ESTHER : Taguchi_2002_J.Biochem_131_801 |
| PubMedSearch : Taguchi_2002_J.Biochem_131_801 |
| PubMedID: 12038975 |
| Gene_locus related to this paper: ralso-PHBC |
| Title : Lipases provide a new mechanistic model for polyhydroxybutyrate (PHB) synthases: characterization of the functional residues in chromatium vinosum PHB synthase - Jia_2000_Biochemistry_39_3927 |
| Author(s) : Jia Y , Kappock TJ , Frick T , Sinskey AJ , Stubbe J |
| Ref : Biochemistry , 39 :3927 , 2000 |
| Abstract : Jia_2000_Biochemistry_39_3927 |
| ESTHER : Jia_2000_Biochemistry_39_3927 |
| PubMedSearch : Jia_2000_Biochemistry_39_3927 |
| PubMedID: 10747780 |
| Title : In vitro polymerization and copolymerization of 3-hydroxypropionyl-CoA with the PHB synthase from Ralstonia eutropha - Song_2000_Biomacromolecules_1_433 |
| Author(s) : Song JJ , Zhang S , Lenz RW , Goodwin S |
| Ref : Biomacromolecules , 1 :433 , 2000 |
| Abstract : Song_2000_Biomacromolecules_1_433 |
| ESTHER : Song_2000_Biomacromolecules_1_433 |
| PubMedSearch : Song_2000_Biomacromolecules_1_433 |
| PubMedID: 11710134 |
| Gene_locus related to this paper: alceu-phbc |
| Title : Cloning, molecular analysis, and expression of the polyhydroxyalkanoic acid synthase (phaC) gene from Chromobacterium violaceum - Kolibachuk_1999_Appl.Environ.Microbiol_65_3561 |
| Author(s) : Kolibachuk D , Miller A , Dennis D |
| Ref : Applied Environmental Microbiology , 65 :3561 , 1999 |
| Abstract : Kolibachuk_1999_Appl.Environ.Microbiol_65_3561 |
| ESTHER : Kolibachuk_1999_Appl.Environ.Microbiol_65_3561 |
| PubMedSearch : Kolibachuk_1999_Appl.Environ.Microbiol_65_3561 |
| PubMedID: 10427049 |
| Gene_locus related to this paper: chrvi-PHAC , vibpa-PHAC |
| Title : PHA synthase from chromatium vinosum: cysteine 149 is involved in covalent catalysis - Muh_1999_Biochemistry_38_826 |
| Author(s) : Muh U , Sinskey AJ , Kirby DP , Lane WS , Stubbe J |
| Ref : Biochemistry , 38 :826 , 1999 |
| Abstract : Muh_1999_Biochemistry_38_826 |
| ESTHER : Muh_1999_Biochemistry_38_826 |
| PubMedSearch : Muh_1999_Biochemistry_38_826 |
| PubMedID: 9888824 |
| Gene_locus related to this paper: chrvi-phbc |