RB3
General
Type : Polymer || Polyhydroxyalkanoate
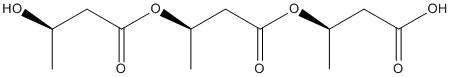
Chemical_Nomenclature : (3R)-3-[(3R)-3-[(3R)-3-hydroxybutanoyl]oxybutanoyl]oxybutanoic acid
Canonical SMILES : CC(CC(=O)OC(C)CC(=O)OC(C)CC(=O)O)O
InChI : InChI=1S\/C12H20O7\/c1-7(13)4-11(16)19-9(3)6-12(17)18-8(2)5-10(14)15\/h7-9,13H,4-6H2,1-3H3,(H,14,15)\/t7-,8-,9-\/m1\/s1
InChIKey : CWLWBMWELZSMPG-IWSPIJDZSA-N
Other name(s) : AC1NUV4R, SCHEMBL804588, ZINC12504507, DB04773, (R)-3-[(R)-3-[(R)-3-Hydroxybutyryloxy]butyryloxy]butyric acid, (3R)-3-[(3R)-3-[(3R)-3-hydroxybutanoyl]oxybutanoyl]oxybutanoic acid
MW : 276.28
Formula : C12H20O7
CAS_number :
PubChem :
UniChem :
Iuphar :
Target
Families : Esterase_phb_PHAZ, PHAZ7_phb_depolymerase
References (3)
| Title : Purification, characterization, and gene cloning of an Aspergillus fumigatus polyhydroxybutyrate depolymerase used for degradation of polyhydroxybutyrate, polyethylene succinate, and polybutylene succinate - Jung_2018_Polym.Degrad.Stab_154_186 |
| Author(s) : Jung HW , Yang MK , Su RC |
| Ref : Polymer Degradation and Stability , 154 :154 , 2018 |
| Abstract : Jung_2018_Polym.Degrad.Stab_154_186 |
| ESTHER : Jung_2018_Polym.Degrad.Stab_154_186 |
| PubMedSearch : Jung_2018_Polym.Degrad.Stab_154_186 |
| PubMedID: |
| Gene_locus related to this paper: aspfu-q4w9v8 |
| Title : Biochemical analysis and structure determination of Paucimonas lemoignei poly(3-hydroxybutyrate) (PHB) depolymerase PhaZ7 muteins reveal the PHB binding site and details of substrate-enzyme interactions - Jendrossek_2013_Mol.Microbiol_90_649 |
| Author(s) : Jendrossek D , Hermawan S , Subedi B , Papageorgiou AC |
| Ref : Molecular Microbiology , 90 :649 , 2013 |
| Abstract : Jendrossek_2013_Mol.Microbiol_90_649 |
| ESTHER : Jendrossek_2013_Mol.Microbiol_90_649 |
| PubMedSearch : Jendrossek_2013_Mol.Microbiol_90_649 |
| PubMedID: 24007310 |
| Gene_locus related to this paper: psele-PHAZ7 |
| Title : The crystal structure of polyhydroxybutyrate depolymerase from Penicillium funiculosum provides insights into the recognition and degradation of biopolyesters - Hisano_2006_J.Mol.Biol_356_993 |
| Author(s) : Hisano T , Kasuya K , Tezuka Y , Ishii N , Kobayashi T , Shiraki M , Oroudjev E , Hansma H , Iwata T , Doi Y , Saito T , Miki K |
| Ref : Journal of Molecular Biology , 356 :993 , 2006 |
| Abstract : Hisano_2006_J.Mol.Biol_356_993 |
| ESTHER : Hisano_2006_J.Mol.Biol_356_993 |
| PubMedSearch : Hisano_2006_J.Mol.Biol_356_993 |
| PubMedID: 16405909 |
| Gene_locus related to this paper: penfu-PHAZ |