Tween-80
General
Type : Polysorbate || Detergent || Oleate
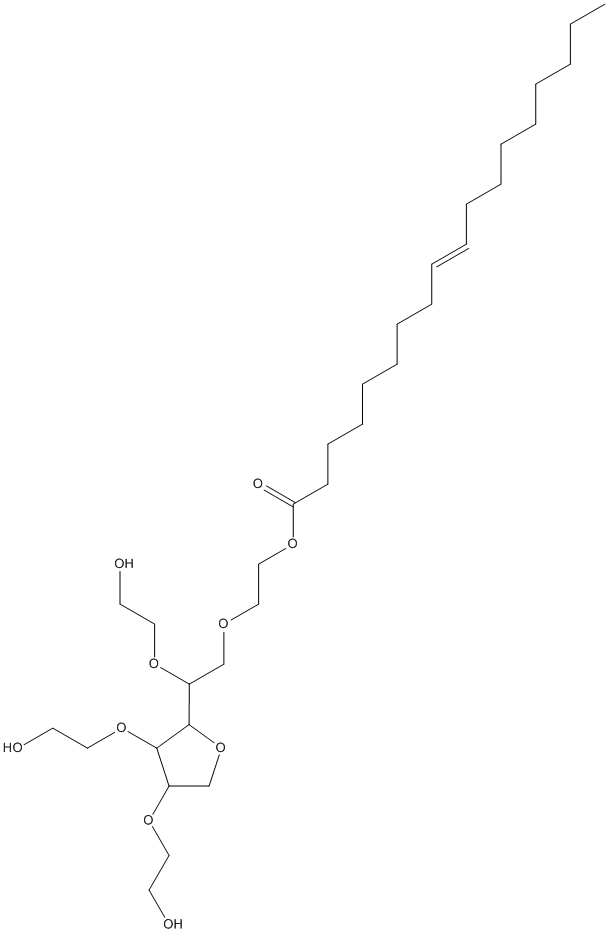
Chemical_Nomenclature : 2-[2-[3,4-bis(2-hydroxyethoxy)oxolan-2-yl]-2-(2-hydroxyethoxy)ethoxy]ethyl (E)-octadec-9-enoate
Canonical SMILES : CCCCCCCCC=CCCCCCCCC(=O)OCCOCC(C1C(C(CO1)OCCO)OCCO)OCCO
InChI : InChI=1S\/C32H60O10\/c1-2-3-4-5-6-7-8-9-10-11-12-13-14-15-16-17-30(36)40-25-24-37-26-28(38-21-18-33)32-31(41-23-20-35)29(27-42-32)39-22-19-34\/h9-10,28-29,31-35H,2-8,11-27H2,1H3\/b10-9+
InChIKey : HDTIFOGXOGLRCB-MDZDMXLPSA-N
Other name(s) : Tween 80, Glycol (polysorbate 80), Polysorbate-81, Polyoxyethylene sorbitan oleate, Polyethylene oxide sorbitan mono-oleate, Polyoxyethylene (20) sorbitan monooleate, AC1NQZCF
MW : 604.82
Formula : C32H60O10
CAS_number :
PubChem :
UniChem :
Iuphar :
Target
Families : Carb_B_Chordata, PAF-Acetylhydrolase, Canar_LipB
References (4)
| Title : Profiling Active Enzymes for Polysorbate Degradation in Biotherapeutics by Activity-Based Protein Profiling - Li_2021_Anal.Chem_93_8161 |
| Author(s) : Li X , Chandra D , Letarte S , Adam GC , Welch J , Yang RS , Rivera S , Bodea S , Dow A , Chi A , Strulson CA , Richardson DD |
| Ref : Analytical Chemistry , 93 :8161 , 2021 |
| Abstract : Li_2021_Anal.Chem_93_8161 |
| ESTHER : Li_2021_Anal.Chem_93_8161 |
| PubMedSearch : Li_2021_Anal.Chem_93_8161 |
| PubMedID: 34032423 |
| Gene_locus related to this paper: human-PLA2G7 |
| Title : Rapid Polysorbate 80 Degradation by Liver Carboxylesterase in a Monoclonal Antibody Formulated Drug Substance at Early Stage Development - Zhang_2020_J.Pharm.Sci_109_3300 |
| Author(s) : Zhang S , Xiao H , Molden R , Qiu H , Li N |
| Ref : J Pharm Sci , 109 :3300 , 2020 |
| Abstract : Zhang_2020_J.Pharm.Sci_109_3300 |
| ESTHER : Zhang_2020_J.Pharm.Sci_109_3300 |
| PubMedSearch : Zhang_2020_J.Pharm.Sci_109_3300 |
| PubMedID: 32721471 |
| Gene_locus related to this paper: crigr-g3i7x7 , crigr-a0a061i7x9 |
| Title : Hydrolysis of Polysorbate 20 and 80 by a Range of Carboxylester Hydrolases - McShan_2016_PDA.J.Pharm.Sci.Technol_70_332 |
| Author(s) : McShan AC , Kei P , Ji JA , Kim DC , Wang YJ |
| Ref : PDA J Pharm Sci Technol , 70 :332 , 2016 |
| Abstract : McShan_2016_PDA.J.Pharm.Sci.Technol_70_332 |
| ESTHER : McShan_2016_PDA.J.Pharm.Sci.Technol_70_332 |
| PubMedSearch : McShan_2016_PDA.J.Pharm.Sci.Technol_70_332 |
| PubMedID: 27020650 |
| Title : Uml2 is a novel CalB-type lipase of Ustilago maydis with phospholipase A activity - Buerth_2014_Appl.Microbiol.Biotechnol_98_4963 |
| Author(s) : Buerth C , Kovacic F , Stock J , Terfruchte M , Wilhelm S , Jaeger KE , Feldbrugge M , Schipper K , Ernst JF , Tielker D |
| Ref : Applied Microbiology & Biotechnology , 98 :4963 , 2014 |
| Abstract : Buerth_2014_Appl.Microbiol.Biotechnol_98_4963 |
| ESTHER : Buerth_2014_Appl.Microbiol.Biotechnol_98_4963 |
| PubMedSearch : Buerth_2014_Appl.Microbiol.Biotechnol_98_4963 |
| PubMedID: 24469105 |
| Gene_locus related to this paper: ustma-q4pep1 |