Ampicillin
General
Type : Antibiotic, Azabicyclo
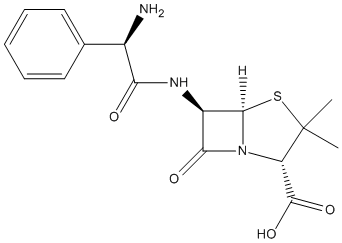
Chemical_Nomenclature : (2S,5R,6R)-6-[[(2R)-2-amino-2-phenylacetyl]amino]-3,3-dimethyl-7-oxo-4-thia-1-azabicyclo[3.2.0]heptane-2-carboxylic acid
Canonical SMILES : CC1(C(N2C(S1)C(C2=O)NC(=O)C(C3=CC=CC=C3)N)C(=O)O)C
InChI : InChI=1S\/C16H19N3O4S\/c1-16(2)11(15(22)23)19-13(21)10(14(19)24-16)18-12(20)9(17)8-6-4-3-5-7-8\/h3-7,9-11,14H,17H2,1-2H3,(H,18,20)(H,22,23)\/t9-,10-,11+,14-\/m1\/s1
InChIKey : AVKUERGKIZMTKX-NJBDSQKTSA-N
Other name(s) : Aminobenzylpenicillin || Ampicillin acid || Amcill || Ampicilline || Polycillin
MW : 349.40
Formula : C16H19N3O4S
CAS_number :
PubChem : 6249
UniChem : AVKUERGKIZMTKX-NJBDSQKTSA-N
Wikipedia : Ampicillin
Target
Structures : 1NX9
Families : Cocaine_esterase, Peptidase_S15, Carb_B_Chordata
References (4)
| Title : Pig liver esterases PLE1 and PLE6: heterologous expression, hydrolysis of common antibiotics and pharmacological consequences - Zhou_2019_Sci.Rep_9_15564 |
| Author(s) : Zhou Q , Xiao Q , Zhang Y , Wang X , Xiao Y , Shi D |
| Ref : Sci Rep , 9 :15564 , 2019 |
| Abstract : |
| PubMedSearch : Zhou_2019_Sci.Rep_9_15564 |
| PubMedID: 31664043 |
| Gene_locus related to this paper: pig-EST1 , pig-a0a1s6l967 |
| Title : Amino ester hydrolase from Xanthomonas campestris pv. campestris, ATCC 33913 for enzymatic synthesis of ampicillin - Blum_2010_J.Mol.Catal.B.Enzym_67_21 |
| Author(s) : Blum JK , Bommarius AS |
| Ref : J Mol Catal B Enzym , 67 :21 , 2010 |
| Abstract : |
| PubMedSearch : Blum_2010_J.Mol.Catal.B.Enzym_67_21 |
| PubMedID: 22087071 |
| Gene_locus related to this paper: xanax-GAA |
| Title : Acetobacter turbidans alpha-amino acid ester hydrolase: how a single mutation improves an antibiotic-producing enzyme - Barends_2006_J.Biol.Chem_281_5804 |
| Author(s) : Barends TR , Polderman-Tijmes JJ , Jekel PA , Williams C , Wybenga G , Janssen DB , Dijkstra BW |
| Ref : Journal of Biological Chemistry , 281 :5804 , 2006 |
| Abstract : |
| PubMedSearch : Barends_2006_J.Biol.Chem_281_5804 |
| PubMedID: 16377627 |
| Gene_locus related to this paper: acepa-AEHA |
| Title : Acetobacter turbidans alpha-amino acid ester hydrolase: merohedral twinning in P21 obscured by pseudo-translational NCS - Barends_2003_Acta.Crystallogr.D.Biol.Crystallogr_59_2237 |
| Author(s) : Barends TR , Dijkstra BW |
| Ref : Acta Crystallographica D Biol Crystallogr , 59 :2237 , 2003 |
| Abstract : |
| PubMedSearch : Barends_2003_Acta.Crystallogr.D.Biol.Crystallogr_59_2237 |
| PubMedID: 14646082 |
| Gene_locus related to this paper: acepa-AEHA |