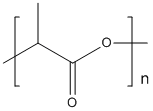
Polylactic-acid
General
Type : Polymer || Polyester || Aliphatic polyester
Chemical_Nomenclature : 2-Hydroxypropanoic acid homopolymer
Canonical SMILES :
InChI :
InChIKey :
Other name(s) : Polylactic acid, poly(L-lactic acid), polylactide, Polylactate, poly(lactic acid), Poly(L-lactide), Poly(D-lactide), PLA, Poly, PLA, PLLA, Luminy L105
References (15)
| Title : Evaluation of the Physical and Shape Memory Properties of Fully Biodegradable Poly(lactic acid) (PLA)\/Poly(butylene adipate terephthalate) (PBAT) Blends - Bianchi_2023_Polymers.(Basel)_15_881 |
| Author(s) : Bianchi M , Dorigato A , Morreale M , Pegoretti A |
| Ref : Polymers (Basel) , 15 :881 , 2023 |
| Abstract : Bianchi_2023_Polymers.(Basel)_15_881 |
| ESTHER : Bianchi_2023_Polymers.(Basel)_15_881 |
| PubMedSearch : Bianchi_2023_Polymers.(Basel)_15_881 |
| PubMedID: 36850164 |
| Title : Toughening Effect of 2,5-Furandicaboxylate Polyesters on Polylactide-Based Renewable Fibers - Fredi_2023_Molecules_28_4811 |
| Author(s) : Fredi G , Zonta E , Dussin A , Bikiaris DN , Papageorgiou GZ , Fambri L , Dorigato A |
| Ref : Molecules , 28 :4811 , 2023 |
| Abstract : Fredi_2023_Molecules_28_4811 |
| ESTHER : Fredi_2023_Molecules_28_4811 |
| PubMedSearch : Fredi_2023_Molecules_28_4811 |
| PubMedID: 37375367 |
| Title : Microbiome dynamics during anaerobic digestion of food waste and the genetic potential for poly (lactic acid) co-digestion - Zhu_2023_Chem.Eng.J__145194 |
| Author(s) : Zhu X , Zhu S , Zhao Z , Kang X , Ju F |
| Ref : Chemical Engineering Journal , :145194 , 2023 |
| Abstract : Zhu_2023_Chem.Eng.J__145194 |
| ESTHER : Zhu_2023_Chem.Eng.J__145194 |
| PubMedSearch : Zhu_2023_Chem.Eng.J__145194 |
| PubMedID: |
| Title : Engineered yeast for the efficient hydrolysis of polylactic acid - Myburgh_2023_Bioresour.Technol__129008 |
| Author(s) : Myburgh MW , Favaro L , van Zyl WH , Viljoen-Bloom M |
| Ref : Bioresour Technol , :129008 , 2023 |
| Abstract : Myburgh_2023_Bioresour.Technol__129008 |
| ESTHER : Myburgh_2023_Bioresour.Technol__129008 |
| PubMedSearch : Myburgh_2023_Bioresour.Technol__129008 |
| PubMedID: 37011843 |
| Gene_locus related to this paper: crysp-Q874E9 |
| Title : Enzymatic hydrolysis of single-use bioplastic items by improved recombinant yeast strains - Myburgh_2023_Bioresour.Technol__129908 |
| Author(s) : Myburgh MW , van Zyl WH , Modesti M , Viljoen-Bloom M , Favaro L |
| Ref : Bioresour Technol , :129908 , 2023 |
| Abstract : Myburgh_2023_Bioresour.Technol__129908 |
| ESTHER : Myburgh_2023_Bioresour.Technol__129908 |
| PubMedSearch : Myburgh_2023_Bioresour.Technol__129908 |
| PubMedID: 37866766 |
| Gene_locus related to this paper: crysp-Q874E9 |
| Title : Biodegradation mechanism of polycaprolactone by a novel esterase MGS0156: a QM\/MM approach - Feng_2020_Environ.Sci.Process.Impacts_22_2332 |
| Author(s) : Feng S , Yue Y , Chen J , Zhou J , Li Y , Zhang Q |
| Ref : Environ Sci Process Impacts , 22 :2332 , 2020 |
| Abstract : Feng_2020_Environ.Sci.Process.Impacts_22_2332 |
| ESTHER : Feng_2020_Environ.Sci.Process.Impacts_22_2332 |
| PubMedSearch : Feng_2020_Environ.Sci.Process.Impacts_22_2332 |
| PubMedID: 33146659 |
| Gene_locus related to this paper: 9zzzz-A0A0G3FEJ8 |
| Title : Structural insights into the unique polylactate-degrading mechanism of Thermobifida alba cutinase - Kitadokoro_2019_FEBS.J_286_2087 |
| Author(s) : Kitadokoro K , Kakara M , Matsui S , Osokoshi R , Thumarat U , Kawai F , Kamitani S |
| Ref : Febs J , 286 :2087 , 2019 |
| Abstract : Kitadokoro_2019_FEBS.J_286_2087 |
| ESTHER : Kitadokoro_2019_FEBS.J_286_2087 |
| PubMedSearch : Kitadokoro_2019_FEBS.J_286_2087 |
| PubMedID: 30761732 |
| Gene_locus related to this paper: 9acto-f7ix06 |
| Title : Screening and Characterization of Novel Polyesterases from Environmental Metagenomes with High Hydrolytic Activity against Synthetic Polyesters - Hajighasemi_2018_Environ.Sci.Technol_52_12388 |
| Author(s) : Hajighasemi M , Tchigvintsev A , Nocek B , Flick R , Popovic A , Hai T , Khusnutdinova AN , Brown G , Xu X , Cui H , Anstett J , Chernikova TN , Bruls T , Le Paslier D , Yakimov MM , Joachimiak A , Golyshina OV , Savchenko A , Golyshin PN , Edwards EA , Yakunin AF |
| Ref : Environ Sci Technol , 52 :12388 , 2018 |
| Abstract : Hajighasemi_2018_Environ.Sci.Technol_52_12388 |
| ESTHER : Hajighasemi_2018_Environ.Sci.Technol_52_12388 |
| PubMedSearch : Hajighasemi_2018_Environ.Sci.Technol_52_12388 |
| PubMedID: 30284819 |
| Gene_locus related to this paper: 9zzzz-a0a0g3fj39 , 9zzzz-a0a0g3fj48 , 9zzzz-A0A0G3FEJ8 , 9bact-a4uz10 |
| Title : Small cause, large effect: Structural characterization of cutinases from Thermobifida cellulosilytica - Ribitsch_2017_Biotechnol.Bioeng_114_2481 |
| Author(s) : Ribitsch D , Hromic A , Zitzenbacher S , Zartl B , Gamerith C , Pellis A , Jungbauer A , Lyskowski A , Steinkellner G , Gruber K , Tscheliessnig R , Herrero Acero E , Guebitz GM |
| Ref : Biotechnol Bioeng , 114 :2481 , 2017 |
| Abstract : Ribitsch_2017_Biotechnol.Bioeng_114_2481 |
| ESTHER : Ribitsch_2017_Biotechnol.Bioeng_114_2481 |
| PubMedSearch : Ribitsch_2017_Biotechnol.Bioeng_114_2481 |
| PubMedID: 28671263 |
| Gene_locus related to this paper: thefu-q6a0i4 , thefu-q6a0i3 |
| Title : Biochemical and Structural Insights into Enzymatic Depolymerization of Polylactic Acid and Other Polyesters by Microbial Carboxylesterases - Hajighasemi_2016_Biomacromolecules_17_2027 |
| Author(s) : Hajighasemi M , Nocek BP , Tchigvintsev A , Brown G , Flick R , Xu X , Cui H , Hai T , Joachimiak A , Golyshin PN , Savchenko A , Edwards EA , Yakunin AF |
| Ref : Biomacromolecules , 17 :2027 , 2016 |
| Abstract : Hajighasemi_2016_Biomacromolecules_17_2027 |
| ESTHER : Hajighasemi_2016_Biomacromolecules_17_2027 |
| PubMedSearch : Hajighasemi_2016_Biomacromolecules_17_2027 |
| PubMedID: 27087107 |
| Gene_locus related to this paper: marav-a1u5n0 , rhopa-q6n9m9 , alcbs-q0vlq1 |
| Title : Tailor-made type II Pseudomonas PHA synthases and their use for the biosynthesis of polylactic acid and its copolymer in recombinant Escherichia coli - Yang_2011_Appl.Microbiol.Biotechnol_90_603 |
| Author(s) : Yang TH , Jung YK , Kang HO , Kim TW , Park SJ , Lee SY |
| Ref : Applied Microbiology & Biotechnology , 90 :603 , 2011 |
| Abstract : Yang_2011_Appl.Microbiol.Biotechnol_90_603 |
| ESTHER : Yang_2011_Appl.Microbiol.Biotechnol_90_603 |
| PubMedSearch : Yang_2011_Appl.Microbiol.Biotechnol_90_603 |
| PubMedID: 21221571 |
| Gene_locus related to this paper: psech-PHAC1 |
| Title : Different enantioselectivity of two types of poly(lactic acid) depolymerases toward poly(l-lactic acid) and poly(d-lactic acid) - Kawai_2011_Polym.Degrad.Stab_96_1342 |
| Author(s) : Kawai F , Nakadai K , Nishioka E , Nakajima H , Ohara H , Masaki K , Iefuji H |
| Ref : Polymer Degradation and Stability , 96 :1342 , 2011 |
| Abstract : Kawai_2011_Polym.Degrad.Stab_96_1342 |
| ESTHER : Kawai_2011_Polym.Degrad.Stab_96_1342 |
| PubMedSearch : Kawai_2011_Polym.Degrad.Stab_96_1342 |
| PubMedID: |
| Gene_locus related to this paper: crysp-Q874E9 |
| Title : Identification and characterization of novel poly(DL-lactic acid) depolymerases from metagenome - Mayumi_2008_Appl.Microbiol.Biotechnol_79_743 |
| Author(s) : Mayumi D , Akutsu-Shigeno Y , Uchiyama H , Nomura N , Nakajima-kambe T |
| Ref : Applied Microbiology & Biotechnology , 79 :743 , 2008 |
| Abstract : Mayumi_2008_Appl.Microbiol.Biotechnol_79_743 |
| ESTHER : Mayumi_2008_Appl.Microbiol.Biotechnol_79_743 |
| PubMedSearch : Mayumi_2008_Appl.Microbiol.Biotechnol_79_743 |
| PubMedID: 18461319 |
| Gene_locus related to this paper: 9bact-a4uz11 , 9bact-a4uz12 , 9bact-a4uz10 , 9bact-a4uz14 |
| Title : Cutinase-like enzyme from the yeast Cryptococcus sp. strain S-2 hydrolyzes polylactic acid and other biodegradable plastics - Masaki_2005_Appl.Environ.Microbiol_71_7548 |
| Author(s) : Masaki K , Kamini NR , Ikeda H , Iefuji H |
| Ref : Applied Environmental Microbiology , 71 :7548 , 2005 |
| Abstract : Masaki_2005_Appl.Environ.Microbiol_71_7548 |
| ESTHER : Masaki_2005_Appl.Environ.Microbiol_71_7548 |
| PubMedSearch : Masaki_2005_Appl.Environ.Microbiol_71_7548 |
| PubMedID: 16269800 |
| Gene_locus related to this paper: crysp-Q874E9 |
| Title : Cloning and sequencing of a poly(DL-lactic acid) depolymerase gene from Paenibacillus amylolyticus strain TB-13 and its functional expression in Escherichia coli - Akutsu-Shigeno_2003_Appl.Environ.Microbiol_69_2498 |
| Author(s) : Akutsu-Shigeno Y , Teeraphatpornchai T , Teamtisong K , Nomura N , Uchiyama H , Nakahara T , Nakajima-kambe T |
| Ref : Applied Environmental Microbiology , 69 :2498 , 2003 |
| Abstract : Akutsu-Shigeno_2003_Appl.Environ.Microbiol_69_2498 |
| ESTHER : Akutsu-Shigeno_2003_Appl.Environ.Microbiol_69_2498 |
| PubMedSearch : Akutsu-Shigeno_2003_Appl.Environ.Microbiol_69_2498 |
| PubMedID: 12732514 |
| Gene_locus related to this paper: paeam-PLAA |