Polycaprolactone
Polycaprolactone (PCL) is a biodegradable polyester. The most common use of polycaprolactone is in the manufacture of speciality polyurethanes. Polycaprolactones impart good resistance to water, oil, solvent and chlorine. PCL is degraded by hydrolysis of its ester linkages in physiological conditions (such as in the human body) and is used as in long-term implants and controlled drug release applications.
General
Type : Polymer, Polyester, Aliphatic polyester
Chemical_Nomenclature : Poly(hexano-6-lactone)
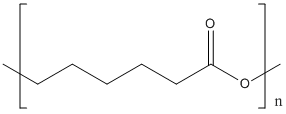
Canonical SMILES :
InChI :
InChIKey :
Other name(s) : PCL || (1,7)-Polyoxepan-2-one || Poly(E''-caprolactone) || Poly(E-caprolactone) || Poly(sigma-caprolactone) || Poly(epsilon-caprolactone) || 2-Oxepanone homopolymer || 6-Caprolactone polymer
Target
Structures : No structure
Families : Cutinase, 6_AlphaBeta_hydrolase, Polyesterase-MGS0156-like, Polyesterase-lipase-cutinase, Bacterial_lip_FamI.1, BD-FAE, Lipase_3
References (69)
| Title : Highly efficient degradation of polybutylene succinate (PBS) and polycaprolactone (PCL) by a recombinant marine fungal cutinase - Lang_2025_Appl.Environ.Microbiol__e0083325 |
| Author(s) : Lang F , Fei F , Sun C , Wu S |
| Ref : Applied Environmental Microbiology , :e0083325 , 2025 |
| Abstract : |
| PubMedSearch : Lang_2025_Appl.Environ.Microbiol__e0083325 |
| PubMedID: 40810542 |
| Gene_locus related to this paper: altal-Aacut10 |
| Title : Biodegradation of Poly(sigma-caprolactone): Microorganisms, Enzymes, and Mechanisms - Krumov_2025_Int.J.Mol.Sci_26_5826 |
| Author(s) : Krumov N , Atanasova N , Boyadzhieva I , Petrov K , Petrova P |
| Ref : Int J Mol Sci , 26 : , 2025 |
| Abstract : |
| PubMedSearch : Krumov_2025_Int.J.Mol.Sci_26_5826 |
| PubMedID: 40565290 |
| Title : Study on Enzymatic Degradation of Polycaprolactone-Based Composite Scaffolds for Tissue Engineering Applications - Buckley_2025_ACS.Appl.Bio.Mater__ |
| Author(s) : Buckley C , Giordano F , Ibrahim R , Parimala Chelvi Ratnamani M , Zhao Y , Wang H |
| Ref : ACS Appl Bio Mater , : , 2025 |
| Abstract : |
| PubMedSearch : Buckley_2025_ACS.Appl.Bio.Mater__ |
| PubMedID: 41037819 |
| Title : Enzymatic biodegradation of Poly(sigma-Caprolactone) (PCL) by a thermostable cutinase from a mesophilic bacteria Mycobacterium marinum - Ravi_2025_Sci.Total.Environ_972_179066 |
| Author(s) : Ravi J , Ponnuraj K , Ragunathan P |
| Ref : Sci Total Environ , 972 :179066 , 2025 |
| Abstract : |
| PubMedSearch : Ravi_2025_Sci.Total.Environ_972_179066 |
| PubMedID: 40088791 |
| Gene_locus related to this paper: mycua-a0pmc3 |
| Title : Enzymatic biodegradation of Poly(sigma-Caprolactone) (PCL) by a thermostable cutinase from a mesophilic bacteria Mycobacterium marinum - Ravi_2025_Sci.Total.Environ_972_179066 |
| Author(s) : Ravi J , Ponnuraj K , Ragunathan P |
| Ref : Sci Total Environ , 972 :179066 , 2025 |
| Abstract : |
| PubMedSearch : Ravi_2025_Sci.Total.Environ_972_179066 |
| PubMedID: 40088791 |
| Gene_locus related to this paper: mycua-a0pmc3 |
| Title : Highly efficient degradation of polybutylene succinate (PBS) and polycaprolactone (PCL) by a recombinant marine fungal cutinase - Lang_2025_Appl.Environ.Microbiol__e0083325 |
| Author(s) : Lang F , Fei F , Sun C , Wu S |
| Ref : Applied Environmental Microbiology , :e0083325 , 2025 |
| Abstract : |
| PubMedSearch : Lang_2025_Appl.Environ.Microbiol__e0083325 |
| PubMedID: 40810542 |
| Gene_locus related to this paper: altal-Aacut10 |
| Title : Highly efficient degradation of polybutylene succinate (PBS) and polycaprolactone (PCL) by a recombinant marine fungal cutinase - Lang_2025_Appl.Environ.Microbiol__e0083325 |
| Author(s) : Lang F , Fei F , Sun C , Wu S |
| Ref : Applied Environmental Microbiology , :e0083325 , 2025 |
| Abstract : |
| PubMedSearch : Lang_2025_Appl.Environ.Microbiol__e0083325 |
| PubMedID: 40810542 |
| Gene_locus related to this paper: altal-Aacut10 |
| Title : Biodegradation of Poly(sigma-caprolactone): Microorganisms, Enzymes, and Mechanisms - Krumov_2025_Int.J.Mol.Sci_26_5826 |
| Author(s) : Krumov N , Atanasova N , Boyadzhieva I , Petrov K , Petrova P |
| Ref : Int J Mol Sci , 26 : , 2025 |
| Abstract : |
| PubMedSearch : Krumov_2025_Int.J.Mol.Sci_26_5826 |
| PubMedID: 40565290 |
| Title : Study on Enzymatic Degradation of Polycaprolactone-Based Composite Scaffolds for Tissue Engineering Applications - Buckley_2025_ACS.Appl.Bio.Mater__ |
| Author(s) : Buckley C , Giordano F , Ibrahim R , Parimala Chelvi Ratnamani M , Zhao Y , Wang H |
| Ref : ACS Appl Bio Mater , : , 2025 |
| Abstract : |
| PubMedSearch : Buckley_2025_ACS.Appl.Bio.Mater__ |
| PubMedID: 41037819 |
| Title : Biodegradation of Poly(sigma-caprolactone): Microorganisms, Enzymes, and Mechanisms - Krumov_2025_Int.J.Mol.Sci_26_5826 |
| Author(s) : Krumov N , Atanasova N , Boyadzhieva I , Petrov K , Petrova P |
| Ref : Int J Mol Sci , 26 : , 2025 |
| Abstract : |
| PubMedSearch : Krumov_2025_Int.J.Mol.Sci_26_5826 |
| PubMedID: 40565290 |
| Title : Study on Enzymatic Degradation of Polycaprolactone-Based Composite Scaffolds for Tissue Engineering Applications - Buckley_2025_ACS.Appl.Bio.Mater__ |
| Author(s) : Buckley C , Giordano F , Ibrahim R , Parimala Chelvi Ratnamani M , Zhao Y , Wang H |
| Ref : ACS Appl Bio Mater , : , 2025 |
| Abstract : |
| PubMedSearch : Buckley_2025_ACS.Appl.Bio.Mater__ |
| PubMedID: 41037819 |
| Title : Degradation and ring-opening polymerization of poly(sigma-caprolactone) by a novel enzyme from Pseudomonas sp. DS0801 - Di_2025_iScience_28_114173 |
| Author(s) : Di Y , Luan W , Sun L , Qi J , Xia H , Li F , Zhai N |
| Ref : iScience , 28 :114173 , 2025 |
| Abstract : |
| PubMedSearch : Di_2025_iScience_28_114173 |
| PubMedID: 41458911 |
| Gene_locus related to this paper: pseae-llipa |
| Title : Discovery of two novel cutinases from a gut yeast of plastic-eating mealworm for polyester depolymerization - Huang_2025_Appl.Environ.Microbiol__e0256224 |
| Author(s) : Huang T , Zhang J , Dong X , Yang Y |
| Ref : Applied Environmental Microbiology , :e0256224 , 2025 |
| Abstract : |
| PubMedSearch : Huang_2025_Appl.Environ.Microbiol__e0256224 |
| PubMedID: 40172219 |
| Gene_locus related to this paper: 9basi-SiCut1 , 9basi-SiCut2 |
| Title : Degradation and ring-opening polymerization of poly(sigma-caprolactone) by a novel enzyme from Pseudomonas sp. DS0801 - Di_2025_iScience_28_114173 |
| Author(s) : Di Y , Luan W , Sun L , Qi J , Xia H , Li F , Zhai N |
| Ref : iScience , 28 :114173 , 2025 |
| Abstract : |
| PubMedSearch : Di_2025_iScience_28_114173 |
| PubMedID: 41458911 |
| Gene_locus related to this paper: pseae-llipa |
| Title : Discovery of two novel cutinases from a gut yeast of plastic-eating mealworm for polyester depolymerization - Huang_2025_Appl.Environ.Microbiol__e0256224 |
| Author(s) : Huang T , Zhang J , Dong X , Yang Y |
| Ref : Applied Environmental Microbiology , :e0256224 , 2025 |
| Abstract : |
| PubMedSearch : Huang_2025_Appl.Environ.Microbiol__e0256224 |
| PubMedID: 40172219 |
| Gene_locus related to this paper: 9basi-SiCut1 , 9basi-SiCut2 |
| Title : Discovery of two novel cutinases from a gut yeast of plastic-eating mealworm for polyester depolymerization - Huang_2025_Appl.Environ.Microbiol__e0256224 |
| Author(s) : Huang T , Zhang J , Dong X , Yang Y |
| Ref : Applied Environmental Microbiology , :e0256224 , 2025 |
| Abstract : |
| PubMedSearch : Huang_2025_Appl.Environ.Microbiol__e0256224 |
| PubMedID: 40172219 |
| Gene_locus related to this paper: 9basi-SiCut1 , 9basi-SiCut2 |
| Title : Enzymatic biodegradation of Poly(sigma-Caprolactone) (PCL) by a thermostable cutinase from a mesophilic bacteria Mycobacterium marinum - Ravi_2025_Sci.Total.Environ_972_179066 |
| Author(s) : Ravi J , Ponnuraj K , Ragunathan P |
| Ref : Sci Total Environ , 972 :179066 , 2025 |
| Abstract : |
| PubMedSearch : Ravi_2025_Sci.Total.Environ_972_179066 |
| PubMedID: 40088791 |
| Gene_locus related to this paper: mycua-a0pmc3 |
| Title : Degradation and ring-opening polymerization of poly(sigma-caprolactone) by a novel enzyme from Pseudomonas sp. DS0801 - Di_2025_iScience_28_114173 |
| Author(s) : Di Y , Luan W , Sun L , Qi J , Xia H , Li F , Zhai N |
| Ref : iScience , 28 :114173 , 2025 |
| Abstract : |
| PubMedSearch : Di_2025_iScience_28_114173 |
| PubMedID: 41458911 |
| Gene_locus related to this paper: pseae-llipa |
| Title : Two Extracellular Poly(sigma-caprolactone)-Degrading Enzymes From Pseudomonas hydrolytica sp. DSWY01(T): Purification, Characterization, and Gene Analysis - Li_2022_Front.Bioeng.Biotechnol_10_835847 |
| Author(s) : Li L , Lin X , Bao J , Xia H , Li F |
| Ref : Front Bioeng Biotechnol , 10 :835847 , 2022 |
| Abstract : |
| PubMedSearch : Li_2022_Front.Bioeng.Biotechnol_10_835847 |
| PubMedID: 35372294 |
| Gene_locus related to this paper: psemy-a4xvf4 , psemy-a4y035 |
| Title : Two Extracellular Poly(sigma-caprolactone)-Degrading Enzymes From Pseudomonas hydrolytica sp. DSWY01(T): Purification, Characterization, and Gene Analysis - Li_2022_Front.Bioeng.Biotechnol_10_835847 |
| Author(s) : Li L , Lin X , Bao J , Xia H , Li F |
| Ref : Front Bioeng Biotechnol , 10 :835847 , 2022 |
| Abstract : |
| PubMedSearch : Li_2022_Front.Bioeng.Biotechnol_10_835847 |
| PubMedID: 35372294 |
| Gene_locus related to this paper: psemy-a4xvf4 , psemy-a4y035 |
| Title : Two Extracellular Poly(sigma-caprolactone)-Degrading Enzymes From Pseudomonas hydrolytica sp. DSWY01(T): Purification, Characterization, and Gene Analysis - Li_2022_Front.Bioeng.Biotechnol_10_835847 |
| Author(s) : Li L , Lin X , Bao J , Xia H , Li F |
| Ref : Front Bioeng Biotechnol , 10 :835847 , 2022 |
| Abstract : |
| PubMedSearch : Li_2022_Front.Bioeng.Biotechnol_10_835847 |
| PubMedID: 35372294 |
| Gene_locus related to this paper: psemy-a4xvf4 , psemy-a4y035 |
| Title : Characterization of Polymer Degrading Lipases, LIP1 and LIP2 From Pseudomonas chlororaphis PA23 - Mohanan_2022_Front.Bioeng.Biotechnol_10_854298 |
| Author(s) : Mohanan N , Wong CH , Budisa N , Levin DB |
| Ref : Front Bioeng Biotechnol , 10 :854298 , 2022 |
| Abstract : |
| PubMedSearch : Mohanan_2022_Front.Bioeng.Biotechnol_10_854298 |
| PubMedID: 35519608 |
| Gene_locus related to this paper: psecl-LIP1 , psecl-LIP2 |
| Title : Characterization of Polymer Degrading Lipases, LIP1 and LIP2 From Pseudomonas chlororaphis PA23 - Mohanan_2022_Front.Bioeng.Biotechnol_10_854298 |
| Author(s) : Mohanan N , Wong CH , Budisa N , Levin DB |
| Ref : Front Bioeng Biotechnol , 10 :854298 , 2022 |
| Abstract : |
| PubMedSearch : Mohanan_2022_Front.Bioeng.Biotechnol_10_854298 |
| PubMedID: 35519608 |
| Gene_locus related to this paper: psecl-LIP1 , psecl-LIP2 |
| Title : Characterization of Polymer Degrading Lipases, LIP1 and LIP2 From Pseudomonas chlororaphis PA23 - Mohanan_2022_Front.Bioeng.Biotechnol_10_854298 |
| Author(s) : Mohanan N , Wong CH , Budisa N , Levin DB |
| Ref : Front Bioeng Biotechnol , 10 :854298 , 2022 |
| Abstract : |
| PubMedSearch : Mohanan_2022_Front.Bioeng.Biotechnol_10_854298 |
| PubMedID: 35519608 |
| Gene_locus related to this paper: psecl-LIP1 , psecl-LIP2 |
| Title : Penicillium fellutanum lipase as a green and ecofriendly biocatalyst for depolymerization of poly (sigma-caprolactone): Biochemical, kinetic, and thermodynamic investigations - Amin_2021_Biotechnol.Appl.Biochem__ |
| Author(s) : Amin M , Bhatti HN , Nawaz S , Bilal M |
| Ref : Biotechnol Appl Biochem , : , 2021 |
| Abstract : |
| PubMedSearch : Amin_2021_Biotechnol.Appl.Biochem__ |
| PubMedID: 33559904 |
| Title : The Bacteroidetes Aequorivita sp. and Kaistella jeonii Produce Promiscuous Esterases With PET-Hydrolyzing Activity - Zhang_2022_Front.Microbiol_12_803896 |
| Author(s) : Zhang H , Perez-Garcia P , Dierkes RF , Applegate V , Schumacher J , Chibani CM , Sternagel S , Preuss L , Weigert S , Schmeisser C , Danso D , Pleiss J , Almeida A , Hocker B , Hallam SJ , Schmitz RA , Smits SHJ , Chow J , Streit WR |
| Ref : Front Microbiol , 12 :803896 , 2021 |
| Abstract : |
| PubMedSearch : Zhang_2022_Front.Microbiol_12_803896 |
| PubMedID: 35069509 |
| Gene_locus related to this paper: flutr-f2ie04 , 9flao-a0a0c1f4u8 , 9flao-kjj39608 , 9flao-a0a330mq60 |
| Title : The Bacteroidetes Aequorivita sp. and Kaistella jeonii Produce Promiscuous Esterases With PET-Hydrolyzing Activity - Zhang_2022_Front.Microbiol_12_803896 |
| Author(s) : Zhang H , Perez-Garcia P , Dierkes RF , Applegate V , Schumacher J , Chibani CM , Sternagel S , Preuss L , Weigert S , Schmeisser C , Danso D , Pleiss J , Almeida A , Hocker B , Hallam SJ , Schmitz RA , Smits SHJ , Chow J , Streit WR |
| Ref : Front Microbiol , 12 :803896 , 2021 |
| Abstract : |
| PubMedSearch : Zhang_2022_Front.Microbiol_12_803896 |
| PubMedID: 35069509 |
| Gene_locus related to this paper: flutr-f2ie04 , 9flao-a0a0c1f4u8 , 9flao-kjj39608 , 9flao-a0a330mq60 |
| Title : The Bacteroidetes Aequorivita sp. and Kaistella jeonii Produce Promiscuous Esterases With PET-Hydrolyzing Activity - Zhang_2022_Front.Microbiol_12_803896 |
| Author(s) : Zhang H , Perez-Garcia P , Dierkes RF , Applegate V , Schumacher J , Chibani CM , Sternagel S , Preuss L , Weigert S , Schmeisser C , Danso D , Pleiss J , Almeida A , Hocker B , Hallam SJ , Schmitz RA , Smits SHJ , Chow J , Streit WR |
| Ref : Front Microbiol , 12 :803896 , 2021 |
| Abstract : |
| PubMedSearch : Zhang_2022_Front.Microbiol_12_803896 |
| PubMedID: 35069509 |
| Gene_locus related to this paper: flutr-f2ie04 , 9flao-a0a0c1f4u8 , 9flao-kjj39608 , 9flao-a0a330mq60 |
| Title : Penicillium fellutanum lipase as a green and ecofriendly biocatalyst for depolymerization of poly (sigma-caprolactone): Biochemical, kinetic, and thermodynamic investigations - Amin_2021_Biotechnol.Appl.Biochem__ |
| Author(s) : Amin M , Bhatti HN , Nawaz S , Bilal M |
| Ref : Biotechnol Appl Biochem , : , 2021 |
| Abstract : |
| PubMedSearch : Amin_2021_Biotechnol.Appl.Biochem__ |
| PubMedID: 33559904 |
| Title : An extracellular lipase from Amycolatopsis mediterannei is a cutinase with plastic degrading activity - Tan_2021_Comput.Struct.Biotechnol.J_19_869 |
| Author(s) : Tan Y , Henehan GT , Kinsella GK , Ryan BJ |
| Ref : Comput Struct Biotechnol J , 19 :869 , 2021 |
| Abstract : |
| PubMedSearch : Tan_2021_Comput.Struct.Biotechnol.J_19_869 |
| PubMedID: 33598102 |
| Gene_locus related to this paper: amymu-a0a0h3des9 |
| Title : Penicillium fellutanum lipase as a green and ecofriendly biocatalyst for depolymerization of poly (sigma-caprolactone): Biochemical, kinetic, and thermodynamic investigations - Amin_2021_Biotechnol.Appl.Biochem__ |
| Author(s) : Amin M , Bhatti HN , Nawaz S , Bilal M |
| Ref : Biotechnol Appl Biochem , : , 2021 |
| Abstract : |
| PubMedSearch : Amin_2021_Biotechnol.Appl.Biochem__ |
| PubMedID: 33559904 |
| Title : An extracellular lipase from Amycolatopsis mediterannei is a cutinase with plastic degrading activity - Tan_2021_Comput.Struct.Biotechnol.J_19_869 |
| Author(s) : Tan Y , Henehan GT , Kinsella GK , Ryan BJ |
| Ref : Comput Struct Biotechnol J , 19 :869 , 2021 |
| Abstract : |
| PubMedSearch : Tan_2021_Comput.Struct.Biotechnol.J_19_869 |
| PubMedID: 33598102 |
| Gene_locus related to this paper: amymu-a0a0h3des9 |
| Title : An extracellular lipase from Amycolatopsis mediterannei is a cutinase with plastic degrading activity - Tan_2021_Comput.Struct.Biotechnol.J_19_869 |
| Author(s) : Tan Y , Henehan GT , Kinsella GK , Ryan BJ |
| Ref : Comput Struct Biotechnol J , 19 :869 , 2021 |
| Abstract : |
| PubMedSearch : Tan_2021_Comput.Struct.Biotechnol.J_19_869 |
| PubMedID: 33598102 |
| Gene_locus related to this paper: amymu-a0a0h3des9 |
| Title : Biodegradation mechanism of polycaprolactone by a novel esterase MGS0156: a QM\/MM approach - Feng_2020_Environ.Sci.Process.Impacts_22_2332 |
| Author(s) : Feng S , Yue Y , Chen J , Zhou J , Li Y , Zhang Q |
| Ref : Environ Sci Process Impacts , 22 :2332 , 2020 |
| Abstract : |
| PubMedSearch : Feng_2020_Environ.Sci.Process.Impacts_22_2332 |
| PubMedID: 33146659 |
| Gene_locus related to this paper: 9zzzz-A0A0G3FEJ8 |
| Title : Biodegradation mechanism of polycaprolactone by a novel esterase MGS0156: a QM\/MM approach - Feng_2020_Environ.Sci.Process.Impacts_22_2332 |
| Author(s) : Feng S , Yue Y , Chen J , Zhou J , Li Y , Zhang Q |
| Ref : Environ Sci Process Impacts , 22 :2332 , 2020 |
| Abstract : |
| PubMedSearch : Feng_2020_Environ.Sci.Process.Impacts_22_2332 |
| PubMedID: 33146659 |
| Gene_locus related to this paper: 9zzzz-A0A0G3FEJ8 |
| Title : Biodegradation mechanism of polycaprolactone by a novel esterase MGS0156: a QM\/MM approach - Feng_2020_Environ.Sci.Process.Impacts_22_2332 |
| Author(s) : Feng S , Yue Y , Chen J , Zhou J , Li Y , Zhang Q |
| Ref : Environ Sci Process Impacts , 22 :2332 , 2020 |
| Abstract : |
| PubMedSearch : Feng_2020_Environ.Sci.Process.Impacts_22_2332 |
| PubMedID: 33146659 |
| Gene_locus related to this paper: 9zzzz-A0A0G3FEJ8 |
| Title : In silico Screening and Heterologous Expression of a Polyethylene Terephthalate Hydrolase (PETase)-Like Enzyme (SM14est) With Polycaprolactone (PCL)-Degrading Activity, From the Marine Sponge-Derived Strain Streptomyces sp. SM14 - Almeida_2019_Front.Microbiol_10_2187 |
| Author(s) : Almeida EL , Rincon AFC , Jackson SA , Dobson ADW , Carrillo Rincon AF |
| Ref : Front Microbiol , 10 :2187 , 2019 |
| Abstract : |
| PubMedSearch : Almeida_2019_Front.Microbiol_10_2187 |
| PubMedID: 31632361 |
| Gene_locus related to this paper: 9actn-SM14est |
| Title : In silico Screening and Heterologous Expression of a Polyethylene Terephthalate Hydrolase (PETase)-Like Enzyme (SM14est) With Polycaprolactone (PCL)-Degrading Activity, From the Marine Sponge-Derived Strain Streptomyces sp. SM14 - Almeida_2019_Front.Microbiol_10_2187 |
| Author(s) : Almeida EL , Rincon AFC , Jackson SA , Dobson ADW , Carrillo Rincon AF |
| Ref : Front Microbiol , 10 :2187 , 2019 |
| Abstract : |
| PubMedSearch : Almeida_2019_Front.Microbiol_10_2187 |
| PubMedID: 31632361 |
| Gene_locus related to this paper: 9actn-SM14est |
| Title : In silico Screening and Heterologous Expression of a Polyethylene Terephthalate Hydrolase (PETase)-Like Enzyme (SM14est) With Polycaprolactone (PCL)-Degrading Activity, From the Marine Sponge-Derived Strain Streptomyces sp. SM14 - Almeida_2019_Front.Microbiol_10_2187 |
| Author(s) : Almeida EL , Rincon AFC , Jackson SA , Dobson ADW , Carrillo Rincon AF |
| Ref : Front Microbiol , 10 :2187 , 2019 |
| Abstract : |
| PubMedSearch : Almeida_2019_Front.Microbiol_10_2187 |
| PubMedID: 31632361 |
| Gene_locus related to this paper: 9actn-SM14est |
| Title : Screening and Characterization of Novel Polyesterases from Environmental Metagenomes with High Hydrolytic Activity against Synthetic Polyesters - Hajighasemi_2018_Environ.Sci.Technol_52_12388 |
| Author(s) : Hajighasemi M , Tchigvintsev A , Nocek B , Flick R , Popovic A , Hai T , Khusnutdinova AN , Brown G , Xu X , Cui H , Anstett J , Chernikova TN , Bruls T , Le Paslier D , Yakimov MM , Joachimiak A , Golyshina OV , Savchenko A , Golyshin PN , Edwards EA , Yakunin AF |
| Ref : Environ Sci Technol , 52 :12388 , 2018 |
| Abstract : |
| PubMedSearch : Hajighasemi_2018_Environ.Sci.Technol_52_12388 |
| PubMedID: 30284819 |
| Gene_locus related to this paper: 9zzzz-a0a0g3fj39 , 9zzzz-a0a0g3fj48 , 9zzzz-A0A0G3FEJ8 , 9bact-a4uz10 |
| Title : Screening and Characterization of Novel Polyesterases from Environmental Metagenomes with High Hydrolytic Activity against Synthetic Polyesters - Hajighasemi_2018_Environ.Sci.Technol_52_12388 |
| Author(s) : Hajighasemi M , Tchigvintsev A , Nocek B , Flick R , Popovic A , Hai T , Khusnutdinova AN , Brown G , Xu X , Cui H , Anstett J , Chernikova TN , Bruls T , Le Paslier D , Yakimov MM , Joachimiak A , Golyshina OV , Savchenko A , Golyshin PN , Edwards EA , Yakunin AF |
| Ref : Environ Sci Technol , 52 :12388 , 2018 |
| Abstract : |
| PubMedSearch : Hajighasemi_2018_Environ.Sci.Technol_52_12388 |
| PubMedID: 30284819 |
| Gene_locus related to this paper: 9zzzz-a0a0g3fj39 , 9zzzz-a0a0g3fj48 , 9zzzz-A0A0G3FEJ8 , 9bact-a4uz10 |
| Title : Screening and Characterization of Novel Polyesterases from Environmental Metagenomes with High Hydrolytic Activity against Synthetic Polyesters - Hajighasemi_2018_Environ.Sci.Technol_52_12388 |
| Author(s) : Hajighasemi M , Tchigvintsev A , Nocek B , Flick R , Popovic A , Hai T , Khusnutdinova AN , Brown G , Xu X , Cui H , Anstett J , Chernikova TN , Bruls T , Le Paslier D , Yakimov MM , Joachimiak A , Golyshina OV , Savchenko A , Golyshin PN , Edwards EA , Yakunin AF |
| Ref : Environ Sci Technol , 52 :12388 , 2018 |
| Abstract : |
| PubMedSearch : Hajighasemi_2018_Environ.Sci.Technol_52_12388 |
| PubMedID: 30284819 |
| Gene_locus related to this paper: 9zzzz-a0a0g3fj39 , 9zzzz-a0a0g3fj48 , 9zzzz-A0A0G3FEJ8 , 9bact-a4uz10 |
| Title : Functional characterization and structural modeling of synthetic polyester-degrading hydrolases from Thermomonospora curvata - Wei_2014_AMB.Express_4_44 |
| Author(s) : Wei R , Oeser T , Then J , Kuhn N , Barth M , Schmidt J , Zimmermann W |
| Ref : AMB Express , 4 :44 , 2014 |
| Abstract : |
| PubMedSearch : Wei_2014_AMB.Express_4_44 |
| PubMedID: 25405080 |
| Gene_locus related to this paper: thecd-d1a9g5 , thecd-d1a2h1 |
| Title : Functional characterization and structural modeling of synthetic polyester-degrading hydrolases from Thermomonospora curvata - Wei_2014_AMB.Express_4_44 |
| Author(s) : Wei R , Oeser T , Then J , Kuhn N , Barth M , Schmidt J , Zimmermann W |
| Ref : AMB Express , 4 :44 , 2014 |
| Abstract : |
| PubMedSearch : Wei_2014_AMB.Express_4_44 |
| PubMedID: 25405080 |
| Gene_locus related to this paper: thecd-d1a9g5 , thecd-d1a2h1 |
| Title : Functional characterization and structural modeling of synthetic polyester-degrading hydrolases from Thermomonospora curvata - Wei_2014_AMB.Express_4_44 |
| Author(s) : Wei R , Oeser T , Then J , Kuhn N , Barth M , Schmidt J , Zimmermann W |
| Ref : AMB Express , 4 :44 , 2014 |
| Abstract : |
| PubMedSearch : Wei_2014_AMB.Express_4_44 |
| PubMedID: 25405080 |
| Gene_locus related to this paper: thecd-d1a9g5 , thecd-d1a2h1 |
| Title : Identification and comparison of cutinases for synthetic polyester degradation - Baker_2012_Appl.Microbiol.Biotechnol_93_229 |
| Author(s) : Baker PJ , Poultney C , Liu Z , Gross R , Montclare JK |
| Ref : Applied Microbiology & Biotechnology , 93 :229 , 2012 |
| Abstract : |
| PubMedSearch : Baker_2012_Appl.Microbiol.Biotechnol_93_229 |
| PubMedID: 21713515 |
| Gene_locus related to this paper: humin-cut |
| Title : Identification and comparison of cutinases for synthetic polyester degradation - Baker_2012_Appl.Microbiol.Biotechnol_93_229 |
| Author(s) : Baker PJ , Poultney C , Liu Z , Gross R , Montclare JK |
| Ref : Applied Microbiology & Biotechnology , 93 :229 , 2012 |
| Abstract : |
| PubMedSearch : Baker_2012_Appl.Microbiol.Biotechnol_93_229 |
| PubMedID: 21713515 |
| Gene_locus related to this paper: humin-cut |
| Title : Identification and comparison of cutinases for synthetic polyester degradation - Baker_2012_Appl.Microbiol.Biotechnol_93_229 |
| Author(s) : Baker PJ , Poultney C , Liu Z , Gross R , Montclare JK |
| Ref : Applied Microbiology & Biotechnology , 93 :229 , 2012 |
| Abstract : |
| PubMedSearch : Baker_2012_Appl.Microbiol.Biotechnol_93_229 |
| PubMedID: 21713515 |
| Gene_locus related to this paper: humin-cut |
| Title : Purification, cloning and expression of an Aspergillus niger lipase for degradation of poly(lactic acid) and poly(e-caprolactone) - Nakajima-Kambe_2012_Polym.Degrad.Stab_97_139 |
| Author(s) : Nakajima-kambe T , Edwinoliver NG , Maeda H , Thirunavukarasu K , Gowthaman MK , Masaki K , Mahalingam S , Kamini NR |
| Ref : Polymer Degradation and Stability , 97 :139 , 2012 |
| Abstract : |
| PubMedSearch : Nakajima-Kambe_2012_Polym.Degrad.Stab_97_139 |
| PubMedID: |
| Gene_locus related to this paper: aspng-g9m5r3 |
| Title : Isolation of a novel cutinase homolog with polyethylene terephthalate-degrading activity from leaf-branch compost by using a metagenomic approach - Sulaiman_2012_Appl.Environ.Microbiol_78_1556 |
| Author(s) : Sulaiman S , Yamato S , Kanaya E , Kim JJ , Koga Y , Takano K , Kanaya S |
| Ref : Applied Environmental Microbiology , 78 :1556 , 2012 |
| Abstract : |
| PubMedSearch : Sulaiman_2012_Appl.Environ.Microbiol_78_1556 |
| PubMedID: 22194294 |
| Gene_locus related to this paper: 9bact-g9by57 |
| Title : Purification, cloning and expression of an Aspergillus niger lipase for degradation of poly(lactic acid) and poly(e-caprolactone) - Nakajima-Kambe_2012_Polym.Degrad.Stab_97_139 |
| Author(s) : Nakajima-kambe T , Edwinoliver NG , Maeda H , Thirunavukarasu K , Gowthaman MK , Masaki K , Mahalingam S , Kamini NR |
| Ref : Polymer Degradation and Stability , 97 :139 , 2012 |
| Abstract : |
| PubMedSearch : Nakajima-Kambe_2012_Polym.Degrad.Stab_97_139 |
| PubMedID: |
| Gene_locus related to this paper: aspng-g9m5r3 |
| Title : Purification, cloning and expression of an Aspergillus niger lipase for degradation of poly(lactic acid) and poly(e-caprolactone) - Nakajima-Kambe_2012_Polym.Degrad.Stab_97_139 |
| Author(s) : Nakajima-kambe T , Edwinoliver NG , Maeda H , Thirunavukarasu K , Gowthaman MK , Masaki K , Mahalingam S , Kamini NR |
| Ref : Polymer Degradation and Stability , 97 :139 , 2012 |
| Abstract : |
| PubMedSearch : Nakajima-Kambe_2012_Polym.Degrad.Stab_97_139 |
| PubMedID: |
| Gene_locus related to this paper: aspng-g9m5r3 |
| Title : Isolation of a novel cutinase homolog with polyethylene terephthalate-degrading activity from leaf-branch compost by using a metagenomic approach - Sulaiman_2012_Appl.Environ.Microbiol_78_1556 |
| Author(s) : Sulaiman S , Yamato S , Kanaya E , Kim JJ , Koga Y , Takano K , Kanaya S |
| Ref : Applied Environmental Microbiology , 78 :1556 , 2012 |
| Abstract : |
| PubMedSearch : Sulaiman_2012_Appl.Environ.Microbiol_78_1556 |
| PubMedID: 22194294 |
| Gene_locus related to this paper: 9bact-g9by57 |
| Title : Isolation of a novel cutinase homolog with polyethylene terephthalate-degrading activity from leaf-branch compost by using a metagenomic approach - Sulaiman_2012_Appl.Environ.Microbiol_78_1556 |
| Author(s) : Sulaiman S , Yamato S , Kanaya E , Kim JJ , Koga Y , Takano K , Kanaya S |
| Ref : Applied Environmental Microbiology , 78 :1556 , 2012 |
| Abstract : |
| PubMedSearch : Sulaiman_2012_Appl.Environ.Microbiol_78_1556 |
| PubMedID: 22194294 |
| Gene_locus related to this paper: 9bact-g9by57 |
| Title : Cutinolytic esterase activity of bacteria isolated from mixed-plant compost and characterization of a cutinase gene from Pseudomonas pseudoalcaligenes - Inglis_2011_Can.J.Microbiol_57_902 |
| Author(s) : Inglis GD , Yanke LJ , Selinger LB |
| Ref : Can J Microbiol , 57 :902 , 2011 |
| Abstract : |
| PubMedSearch : Inglis_2011_Can.J.Microbiol_57_902 |
| PubMedID: 22029433 |
| Gene_locus related to this paper: pseol-e9kjl1 |
| Title : Cutinolytic esterase activity of bacteria isolated from mixed-plant compost and characterization of a cutinase gene from Pseudomonas pseudoalcaligenes - Inglis_2011_Can.J.Microbiol_57_902 |
| Author(s) : Inglis GD , Yanke LJ , Selinger LB |
| Ref : Can J Microbiol , 57 :902 , 2011 |
| Abstract : |
| PubMedSearch : Inglis_2011_Can.J.Microbiol_57_902 |
| PubMedID: 22029433 |
| Gene_locus related to this paper: pseol-e9kjl1 |
| Title : Cutinolytic esterase activity of bacteria isolated from mixed-plant compost and characterization of a cutinase gene from Pseudomonas pseudoalcaligenes - Inglis_2011_Can.J.Microbiol_57_902 |
| Author(s) : Inglis GD , Yanke LJ , Selinger LB |
| Ref : Can J Microbiol , 57 :902 , 2011 |
| Abstract : |
| PubMedSearch : Inglis_2011_Can.J.Microbiol_57_902 |
| PubMedID: 22029433 |
| Gene_locus related to this paper: pseol-e9kjl1 |
| Title : Enzymatic surface hydrolysis of poly(ethylene terephthalate) and bis(benzoyloxyethyl) terephthalate by lipase and cutinase in the presence of surface active molecules - Eberl_2009_J.Biotechnol_143_207 |
| Author(s) : Eberl A , Heumann S , Bruckner T , Araujo R , Cavaco-Paulo A , Kaufmann F , Kroutil W , Guebitz GM |
| Ref : J Biotechnol , 143 :207 , 2009 |
| Abstract : |
| PubMedSearch : Eberl_2009_J.Biotechnol_143_207 |
| PubMedID: 19616594 |
| Title : Enzymatic surface hydrolysis of poly(ethylene terephthalate) and bis(benzoyloxyethyl) terephthalate by lipase and cutinase in the presence of surface active molecules - Eberl_2009_J.Biotechnol_143_207 |
| Author(s) : Eberl A , Heumann S , Bruckner T , Araujo R , Cavaco-Paulo A , Kaufmann F , Kroutil W , Guebitz GM |
| Ref : J Biotechnol , 143 :207 , 2009 |
| Abstract : |
| PubMedSearch : Eberl_2009_J.Biotechnol_143_207 |
| PubMedID: 19616594 |
| Title : Structural and functional studies of Aspergillus oryzae cutinase: enhanced thermostability and hydrolytic activity of synthetic ester and polyester degradation - Liu_2009_J.Am.Chem.Soc_131_15711 |
| Author(s) : Liu Z , Gosser Y , Baker PJ , Ravee Y , Lu Z , Alemu G , Li H , Butterfoss GL , Kong XP , Gross R , Montclare JK |
| Ref : Journal of the American Chemical Society , 131 :15711 , 2009 |
| Abstract : |
| PubMedSearch : Liu_2009_J.Am.Chem.Soc_131_15711 |
| PubMedID: 19810726 |
| Gene_locus related to this paper: aspor-cutas |
| Title : Structural and functional studies of Aspergillus oryzae cutinase: enhanced thermostability and hydrolytic activity of synthetic ester and polyester degradation - Liu_2009_J.Am.Chem.Soc_131_15711 |
| Author(s) : Liu Z , Gosser Y , Baker PJ , Ravee Y , Lu Z , Alemu G , Li H , Butterfoss GL , Kong XP , Gross R , Montclare JK |
| Ref : Journal of the American Chemical Society , 131 :15711 , 2009 |
| Abstract : |
| PubMedSearch : Liu_2009_J.Am.Chem.Soc_131_15711 |
| PubMedID: 19810726 |
| Gene_locus related to this paper: aspor-cutas |
| Title : Structural and functional studies of Aspergillus oryzae cutinase: enhanced thermostability and hydrolytic activity of synthetic ester and polyester degradation - Liu_2009_J.Am.Chem.Soc_131_15711 |
| Author(s) : Liu Z , Gosser Y , Baker PJ , Ravee Y , Lu Z , Alemu G , Li H , Butterfoss GL , Kong XP , Gross R , Montclare JK |
| Ref : Journal of the American Chemical Society , 131 :15711 , 2009 |
| Abstract : |
| PubMedSearch : Liu_2009_J.Am.Chem.Soc_131_15711 |
| PubMedID: 19810726 |
| Gene_locus related to this paper: aspor-cutas |
| Title : Enzymatic surface hydrolysis of poly(ethylene terephthalate) and bis(benzoyloxyethyl) terephthalate by lipase and cutinase in the presence of surface active molecules - Eberl_2009_J.Biotechnol_143_207 |
| Author(s) : Eberl A , Heumann S , Bruckner T , Araujo R , Cavaco-Paulo A , Kaufmann F , Kroutil W , Guebitz GM |
| Ref : J Biotechnol , 143 :207 , 2009 |
| Abstract : |
| PubMedSearch : Eberl_2009_J.Biotechnol_143_207 |
| PubMedID: 19616594 |
| Title : Cutinase-like enzyme from the yeast Cryptococcus sp. strain S-2 hydrolyzes polylactic acid and other biodegradable plastics - Masaki_2005_Appl.Environ.Microbiol_71_7548 |
| Author(s) : Masaki K , Kamini NR , Ikeda H , Iefuji H |
| Ref : Applied Environmental Microbiology , 71 :7548 , 2005 |
| Abstract : |
| PubMedSearch : Masaki_2005_Appl.Environ.Microbiol_71_7548 |
| PubMedID: 16269800 |
| Gene_locus related to this paper: crysp-Q874E9 |
| Title : Cutinase-like enzyme from the yeast Cryptococcus sp. strain S-2 hydrolyzes polylactic acid and other biodegradable plastics - Masaki_2005_Appl.Environ.Microbiol_71_7548 |
| Author(s) : Masaki K , Kamini NR , Ikeda H , Iefuji H |
| Ref : Applied Environmental Microbiology , 71 :7548 , 2005 |
| Abstract : |
| PubMedSearch : Masaki_2005_Appl.Environ.Microbiol_71_7548 |
| PubMedID: 16269800 |
| Gene_locus related to this paper: crysp-Q874E9 |
| Title : Cutinase-like enzyme from the yeast Cryptococcus sp. strain S-2 hydrolyzes polylactic acid and other biodegradable plastics - Masaki_2005_Appl.Environ.Microbiol_71_7548 |
| Author(s) : Masaki K , Kamini NR , Ikeda H , Iefuji H |
| Ref : Applied Environmental Microbiology , 71 :7548 , 2005 |
| Abstract : |
| PubMedSearch : Masaki_2005_Appl.Environ.Microbiol_71_7548 |
| PubMedID: 16269800 |
| Gene_locus related to this paper: crysp-Q874E9 |
| Title : Fusarium polycaprolactone depolymerase is cutinase - Murphy_1996_Appl.Environ.Microbiol_62_456 |
| Author(s) : Murphy CA , Cameron JA , Huang SJ , Vinopal RT |
| Ref : Applied Environmental Microbiology , 62 :456 , 1996 |
| Abstract : |
| PubMedSearch : Murphy_1996_Appl.Environ.Microbiol_62_456 |
| PubMedID: 8593048 |
| Gene_locus related to this paper: fusso-CUT2 , fusso-CUT3 |
| Title : Fusarium polycaprolactone depolymerase is cutinase - Murphy_1996_Appl.Environ.Microbiol_62_456 |
| Author(s) : Murphy CA , Cameron JA , Huang SJ , Vinopal RT |
| Ref : Applied Environmental Microbiology , 62 :456 , 1996 |
| Abstract : |
| PubMedSearch : Murphy_1996_Appl.Environ.Microbiol_62_456 |
| PubMedID: 8593048 |
| Gene_locus related to this paper: fusso-CUT2 , fusso-CUT3 |
| Title : Fusarium polycaprolactone depolymerase is cutinase - Murphy_1996_Appl.Environ.Microbiol_62_456 |
| Author(s) : Murphy CA , Cameron JA , Huang SJ , Vinopal RT |
| Ref : Applied Environmental Microbiology , 62 :456 , 1996 |
| Abstract : |
| PubMedSearch : Murphy_1996_Appl.Environ.Microbiol_62_456 |
| PubMedID: 8593048 |
| Gene_locus related to this paper: fusso-CUT2 , fusso-CUT3 |