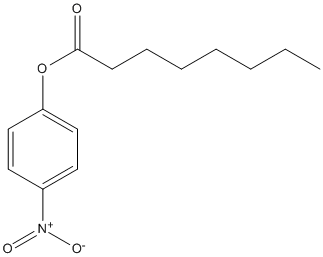
Paranitrophenyl-octanoate
General
Type : Water-insoluble medium and long chain ester || pNP || Chromogen || Octanoate
Chemical_Nomenclature : (4-nitrophenyl) octanoate
Canonical SMILES : CCCCCCCC(=O)OC1=CC=C(C=C1)[N+](=O)[O-]
InChI : InChI=1S\/C14H19NO4\/c1-2-3-4-5-6-7-14(16)19-13-10-8-12(9-11-13)15(17)18\/h8-11H,2-7H2,1H3
InChIKey : GGIDEJQGAZSTES-UHFFFAOYSA-N
Other name(s) : pNP-C8, p-nitrophenyl octanoate, 4-Nitrophenyl octanoate, 4-Nitrophenyl caprylate, 4-NPO, p-NP caprylate, pNP-caprylate, pNP caprylate, p-NP octanoate, PNPC, p-NPC, pNP8, pNP-octanoate, P-Nitrophenyl caprylate, P-Nitrophenylcaprylate, Octanoic acid, 4-nitrophenyl ester, Caprylic acid 4-nitrophenyl ester
Target
Families : Bacterial_lip_FamI.5, CarbLipBact_2, GTSAGmotif, Bact_LipEH166_FamXII, Bacterial_EstLip_FamX, Lipase_2, Pectinacetylesterase-Notum, Bacterial_lip_FamI.6, 6_AlphaBeta_hydrolase, Fungal_carboxylesterase_lipase, ACPH_Peptidase_S9, Hormone-sensitive_lipase_like, LYsophospholipase_carboxylesterase, AlphaBeta_hydrolase, ABHD6-Lip, Cutinase, Polyesterase-lipase-cutinase, Lipase_3, FAE-Bacterial-promiscuous
References (15)
| Title : Structure-guided rational design of the Geobacillus thermoglucosidasius feruloyl esterase GthFAE to improve its thermostability - Yang_2022_Biochem.Biophys.Res.Commun_600_117 |
| Author(s) : Yang W , Sun L , Dong P , Chen Y , Zhang H , Huang X , Wu L , Chen L , Jing D , Wu Y |
| Ref : Biochemical & Biophysical Research Communications , 600 :117 , 2022 |
| Abstract : Yang_2022_Biochem.Biophys.Res.Commun_600_117 |
| ESTHER : Yang_2022_Biochem.Biophys.Res.Commun_600_117 |
| PubMedSearch : Yang_2022_Biochem.Biophys.Res.Commun_600_117 |
| PubMedID: 35219099 |
| Gene_locus related to this paper: partm-GthFAE |
| Title : Trypanosoma brucei Acyl-Protein Thioesterase-like (TbAPT-L) Is a Lipase with Esterase Activity for Short and Medium-Chain Fatty Acids but Has No Depalmitoylation Activity - Brown_2022_Pathogens_11_1245 |
| Author(s) : Brown RWB , Sharma AI , Villanueva MR , Li X , Onguka O , Zilbermintz L , Nguyen H , Falk BA , Olson CL , Taylor JM , Epting CL , Kathayat RS , Amara N , Dickinson BC , Bogyo M , Engman DM |
| Ref : Pathogens , 11 : , 2022 |
| Abstract : Brown_2022_Pathogens_11_1245 |
| ESTHER : Brown_2022_Pathogens_11_1245 |
| PubMedSearch : Brown_2022_Pathogens_11_1245 |
| PubMedID: 36364996 |
| Gene_locus related to this paper: tryb2-q7yxi0 |
| Title : Molecular and functional basis of a novel Amazonian Dark Earth Esterase 1 (Ade1) with hysteresis behavior and quorum-quenching activity - Vinces_2021_Biorxiv__ |
| Author(s) : Vinces TC , de Souza AS , Carvalho CF , Teixeira RD , Bismara BAP , Vicente EJ , Pereira JO , de Souza RF , Yonamine M , Marana SR , Farah CS , Guzzo CR |
| Ref : Biorxiv , : , 2021 |
| Abstract : Vinces_2021_Biorxiv__ |
| ESTHER : Vinces_2021_Biorxiv__ |
| PubMedSearch : Vinces_2021_Biorxiv__ |
| PubMedID: |
| Gene_locus related to this paper: 9bact-6EB3 |
| Title : The Bacteroidetes Aequorivita sp. and Kaistella jeonii Produce Promiscuous Esterases With PET-Hydrolyzing Activity - Zhang_2022_Front.Microbiol_12_803896 |
| Author(s) : Zhang H , Perez-Garcia P , Dierkes RF , Applegate V , Schumacher J , Chibani CM , Sternagel S , Preuss L , Weigert S , Schmeisser C , Danso D , Pleiss J , Almeida A , Hocker B , Hallam SJ , Schmitz RA , Smits SHJ , Chow J , Streit WR |
| Ref : Front Microbiol , 12 :803896 , 2021 |
| Abstract : Zhang_2022_Front.Microbiol_12_803896 |
| ESTHER : Zhang_2022_Front.Microbiol_12_803896 |
| PubMedSearch : Zhang_2022_Front.Microbiol_12_803896 |
| PubMedID: 35069509 |
| Gene_locus related to this paper: flutr-f2ie04 , 9flao-a0a0c1f4u8 , 9flao-kjj39608 , 9flao-a0a330mq60 |
| Title : Phylogenetic analysis and in-depth characterization of functionally and structurally diverse CE5 cutinases - Novy_2021_J.Biol.Chem__101302 |
| Author(s) : Novy V , Carneiro LV , Shin JH , Larsbrink J , Olsson L |
| Ref : Journal of Biological Chemistry , :101302 , 2021 |
| Abstract : Novy_2021_J.Biol.Chem__101302 |
| ESTHER : Novy_2021_J.Biol.Chem__101302 |
| PubMedSearch : Novy_2021_J.Biol.Chem__101302 |
| PubMedID: 34653507 |
| Gene_locus related to this paper: 9pezi-s4vch4 , aspfu-q4x1n0 , crysp-Q874E9 , emeni-q5b2c1 , fusso-cutas , pyrbr-Q9Y7G8 , copci-b9u443 , thite-g2rae6 , strsw-c9zcr8 |
| Title : Pseudomonas aeruginosa esterase PA2949, a bacterial homolog of the human membrane esterase ABHD6: expression, purification and crystallization - Bleffert_2019_Acta.Crystallogr.F.Struct.Biol.Commun_75_270 |
| Author(s) : Bleffert F , Granzin J , Gohlke H , Batra-Safferling R , Jaeger KE , Kovacic F |
| Ref : Acta Crystallographica F Struct Biol Commun , 75 :270 , 2019 |
| Abstract : Bleffert_2019_Acta.Crystallogr.F.Struct.Biol.Commun_75_270 |
| ESTHER : Bleffert_2019_Acta.Crystallogr.F.Struct.Biol.Commun_75_270 |
| PubMedSearch : Bleffert_2019_Acta.Crystallogr.F.Struct.Biol.Commun_75_270 |
| PubMedID: 30950828 |
| Gene_locus related to this paper: pseae-PA2949 |
| Title : Understanding the interactions of different substrates with wild-type and mutant acylaminoacyl peptidase using molecular dynamics simulations - Zhu_2017_J.Biomol.Struct.Dyn__1 |
| Author(s) : Zhu J , Wang Y , Li X , Han W , Zhao L |
| Ref : J Biomol Struct Dyn , :1 , 2017 |
| Abstract : Zhu_2017_J.Biomol.Struct.Dyn__1 |
| ESTHER : Zhu_2017_J.Biomol.Struct.Dyn__1 |
| PubMedSearch : Zhu_2017_J.Biomol.Struct.Dyn__1 |
| PubMedID: 29235404 |
| Gene_locus related to this paper: aerpe-APE1547 |
| Title : Characterization of a poly(butylene adipate-co-terephthalate)-hydrolyzing lipase from Pelosinus fermentans - Biundo_2016_Appl.Microbiol.Biotechnol_100_1753 |
| Author(s) : Biundo A , Hromic A , Pavkov-Keller T , Gruber K , Quartinello F , Haernvall K , Perz V , Arrell MS , Zinn M , Ribitsch D , Guebitz GM |
| Ref : Applied Microbiology & Biotechnology , 100 :1753 , 2016 |
| Abstract : Biundo_2016_Appl.Microbiol.Biotechnol_100_1753 |
| ESTHER : Biundo_2016_Appl.Microbiol.Biotechnol_100_1753 |
| PubMedSearch : Biundo_2016_Appl.Microbiol.Biotechnol_100_1753 |
| PubMedID: 26490551 |
| Gene_locus related to this paper: 9firm-a0a0a0ymq9 |
| Title : Directed evolution of new and improved enzyme functions using an evolutionary intermediate and multidirectional search - Porter_2015_ACS.Chem.Biol_10_611 |
| Author(s) : Porter JL , Boon PL , Murray TP , Huber T , Collyer CA , Ollis DL |
| Ref : ACS Chemical Biology , 10 :611 , 2015 |
| Abstract : Porter_2015_ACS.Chem.Biol_10_611 |
| ESTHER : Porter_2015_ACS.Chem.Biol_10_611 |
| PubMedSearch : Porter_2015_ACS.Chem.Biol_10_611 |
| PubMedID: 25419863 |
| Gene_locus related to this paper: psepu-clcd1 |
| Title : Structural insights into enzymatic degradation of oxidized polyvinyl alcohol - Yang_2014_Chembiochem_15_1882 |
| Author(s) : Yang Y , Ko TP , Liu L , Li J , Huang CH , Chan HC , Ren F , Jia D , Wang AH , Guo RT , Chen J , Du G |
| Ref : Chembiochem , 15 :1882 , 2014 |
| Abstract : Yang_2014_Chembiochem_15_1882 |
| ESTHER : Yang_2014_Chembiochem_15_1882 |
| PubMedSearch : Yang_2014_Chembiochem_15_1882 |
| PubMedID: 25044912 |
| Gene_locus related to this paper: psesp-OPH , sphs1-OPH |
| Title : A novel thermostable carboxylesterase from Geobacillus thermodenitrificans: evidence for a new carboxylesterase family - Charbonneau_2010_J.Biochem_148_299 |
| Author(s) : Charbonneau DM , Meddeb-Mouelhi F , Beauregard M |
| Ref : J Biochem , 148 :299 , 2010 |
| Abstract : Charbonneau_2010_J.Biochem_148_299 |
| ESTHER : Charbonneau_2010_J.Biochem_148_299 |
| PubMedSearch : Charbonneau_2010_J.Biochem_148_299 |
| PubMedID: 20587647 |
| Gene_locus related to this paper: geotd-g3jwz2 |
| Title : Morphing activity between structurally similar enzymes: from heme-free bromoperoxidase to lipase - Chen_2009_Biochemistry_48_11496 |
| Author(s) : Chen B , Cai Z , Wu W , Huang Y , Pleiss J , Lin Z |
| Ref : Biochemistry , 48 :11496 , 2009 |
| Abstract : Chen_2009_Biochemistry_48_11496 |
| ESTHER : Chen_2009_Biochemistry_48_11496 |
| PubMedSearch : Chen_2009_Biochemistry_48_11496 |
| PubMedID: 19883129 |
| Title : Characterization and heterologous gene expression of a novel esterase from Lactobacillus casei CL96 - Choi_2004_Appl.Environ.Microbiol_70_3213 |
| Author(s) : Choi YJ , Miguez CB , Lee BH |
| Ref : Applied Environmental Microbiology , 70 :3213 , 2004 |
| Abstract : Choi_2004_Appl.Environ.Microbiol_70_3213 |
| ESTHER : Choi_2004_Appl.Environ.Microbiol_70_3213 |
| PubMedSearch : Choi_2004_Appl.Environ.Microbiol_70_3213 |
| PubMedID: 15184114 |
| Gene_locus related to this paper: lacca-ESTI |
| Title : A novel extracellular esterase from Bacillus subtilis and its conversion to a monoacylglycerol hydrolase - Eggert_2000_Eur.J.Biochem_267_6459 |
| Author(s) : Eggert T , Pencreac'h G , Douchet I , Verger R , Jaeger KE |
| Ref : European Journal of Biochemistry , 267 :6459 , 2000 |
| Abstract : Eggert_2000_Eur.J.Biochem_267_6459 |
| ESTHER : Eggert_2000_Eur.J.Biochem_267_6459 |
| PubMedSearch : Eggert_2000_Eur.J.Biochem_267_6459 |
| PubMedID: 11029590 |
| Gene_locus related to this paper: bacsu-LIPB |
| Title : Purification and partial characterization of a novel thermophilic carboxylesterase with high mesophilic specific activity - Wood_1995_Enzyme.Microb.Technol_17_816 |
| Author(s) : Wood AN , Fernandez-Lafuente R , Cowan DA |
| Ref : Enzyme Microb Technol , 17 :816 , 1995 |
| Abstract : Wood_1995_Enzyme.Microb.Technol_17_816 |
| ESTHER : Wood_1995_Enzyme.Microb.Technol_17_816 |
| PubMedSearch : Wood_1995_Enzyme.Microb.Technol_17_816 |
| PubMedID: 7576531 |
| Gene_locus related to this paper: geost-est50 |